A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription
- PMID: 12727881
- PMCID: PMC156091
- DOI: 10.1093/emboj/cdg219
A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription
Abstract
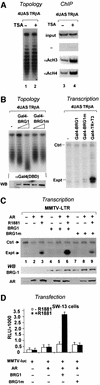
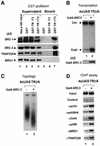
Transcriptional activation from chromatin by nuclear receptors (NRs) requires multiple cofactors including CBP/p300, SWI/SNF and Mediator. How NRs recruit these multiple cofactors is not clear. Here we show that activation by androgen receptor and thyroid hormone receptor is associated with the promoter targeting of SRC family members, p300, SWI/SNF and the Mediator complex. We show that recruitment of SWI/SNF leads to chromatin remodeling with altered DNA topology, and that both SWI/SNF and p300 histone acetylase activity are required for hormone-dependent activation. Importantly, we show that both the SWI/SNF and Mediator complexes can be targeted to chromatin by p300, which itself is recruited through interaction with SRC coactivators. Furthermore, histone acetylation by CBP/p300 facilitates the recruitment of SWI/SNF and Mediator. Thus, our data indicate that multiple cofactors required for activation are not all recruited through their direct interactions with NRs and underscore a role of cofactor-cofactor interaction and histone modification in coordinating the recruitment of multiple cofactors.
Figures





References
-
- Aalfs J.D., Narlikar,G.J. and Kingston,R.E. (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem., 276, 34270–34278. - PubMed
-
- Agalioti T., Chen,G. and Thanos,D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381–392. - PubMed
-
- Becker P.B. and Horz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

