Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse
- PMID: 12727891
- PMCID: PMC156078
- DOI: 10.1093/emboj/cdg206
Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse
Abstract
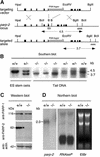
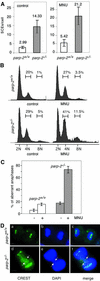
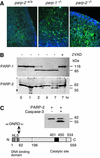
The DNA damage-dependent poly(ADP-ribose) polymerases, PARP-1 and PARP-2, homo- and heterodimerize and are both involved in the base excision repair (BER) pathway. Here, we report that mice carrying a targeted disruption of the PARP-2 gene are sensitive to ionizing radiation. Following alkylating agent treatment, parp-2(-/-)-derived mouse embryonic fibroblasts exhibit increased post-replicative genomic instability, G(2)/M accumulation and chromosome mis-segregation accompanying kinetochore defects. Moreover, parp-1(-/-)parp-2(-/-) double mutant mice are not viable and die at the onset of gastrulation, demonstrating that the expression of both PARP-1 and PARP-2 and/or DNA-dependent poly(ADP-ribosyl) ation is essential during early embryogenesis. Interestingly, specific female embryonic lethality is observed in parp-1(+/-)parp-2(-/-) mutants at E9.5. Meta phase analyses of E8.5 embryonic fibroblasts highlight a specific instability of the X chromosome in those females, but not in males. Together, these results support the notion that PARP-1 and PARP-2 possess both overlapping and non-redundant functions in the maintenance of genomic stability.
Figures



References
-
- Allen J.W., Shuler,C.F., Mendes,R.W. and Latt,S.A. (1977) A simplified technique for in vivo analysis of sister-chromatid exchanges using 5-bromodeoxyuridine tablets. Cytogenet. Cell Genet., 18, 231–237. - PubMed
-
- Amé J.C. et al. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. - PubMed
-
- Amé J.C., Schreiber,V., Fraulob,V., Dollé,P., de Murcia,G. and Niedergang,C.P. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J. Biol. Chem., 276, 11092–11099. - PubMed
-
- Avner P. and Heard,E. (2001) X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet., 2, 59–67. - PubMed
-
- Benchoua A., Couriaud,C., Guegan,C., Tartier,L., Couvert,P., Friocourt,G., Chelly,J., Menissier-de Murcia,J. and Onteniente,B. (2002) Active caspase-8 translocates into the nucleus of apoptotic cells to inactivate poly(ADP-ribose) polymerase-2. J. Biol. Chem., 277, 34217–34222. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

