Retroposed copies of the HMG genes: a window to genome dynamics
- PMID: 12727900
- PMCID: PMC430908
- DOI: 10.1101/gr.893803
Retroposed copies of the HMG genes: a window to genome dynamics
Abstract
Retroposed copies (RPCs) of genes are functional (intronless paralogs) or nonfunctional (processed pseudogenes) copies derived from mRNA through a process of retrotransposition. Previous studies found that gene families involved in mRNA translation or nuclear function were more likely to have large numbers of RPCs. Here we characterize RPCs of the few families coding for the abundant high-mobility-group (HMG) proteins in humans. Using an algorithm we developed, we identified and studied 219 HMG RPCs. For slightly more than 10% of these RPCs, we found evidence indicating expression. Furthermore, eight of these are potentially new members of the HMG families of proteins. For three RPCs, the evidence indicated expression as part of other transcripts; in all of these, we found the presence of alternative splicing or multiple polyadenylation signals. RPC distribution among the HMGs was not even, with 33-65 each for HMGB1, HMGB3, HMGN1, and HMGN2, and 0-6 each for HMGA1, HMGA2, HMGB2, and HMGN3. Analysis of the sequences flanking the RPCs revealed that the junction between the target site duplications and the 5'-flanking sequences exhibited the same TT/AAAA consensus found for the L1 endonuclease, supporting an L1-mediated retrotransposition mechanism. Finally, because our algorithm included aligning RPC flanking sequences with the corresponding HMG genomic sequence, we were able to identify transcribed regions of HMG genes that were not part of the published mRNA sequences.
Figures
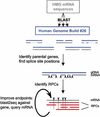





References
-
- Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Birger Y., Ito, Y., West, K.L., Landsman, D., and Bustin, M. 2001. HMGN4, a newly discovered nucleosome-binding protein encoded by an intronless gene. DNA Cell. Biol. 20: 257-264. - PubMed
-
- Boeke J.D. 1997. LINEs and Alus—The polyA connection. Nat. Genet. 16: 6-7. - PubMed
-
- Dunham I., Shimizu, N., Roe, B.A., Chissoe, S., Hunt, A.R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., et al. 1999. The DNA sequence of human Chromosome 22. Nature 402: 489-495. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
