Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes
- PMID: 12727904
- PMCID: PMC430942
- DOI: 10.1101/gr.1152803
Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes
Abstract
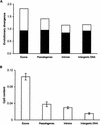
Point mutation rates in exons (synonymous sites) and noncoding (introns and intergenic) regions are generally assumed to be the same. However, comparative sequence analyses of synonymous substitutions in exons (81 genes) and that of long intergenic fragments (141.3 kbp) of human and chimpanzee genomes reveal a 30%-60% higher mutation rate in exons than in noncoding DNA. We propose a differential CpG content hypothesis to explain this fundamental, and seemingly unintuitive, pattern. We find that the increased exonic rate is the result of the relative overabundance of synonymous sites involved in CpG dinucleotides, as the evolutionary divergence in non-CpG sites is similar in noncoding DNA and synonymous sites of exons. Expectations and predictions of our hypothesis are confirmed in comparisons involving more distantly related species, including human-orangutan, human-baboon, and human-macaque. Our results suggest an underlying mechanism for higher mutation rate in GC-rich genomic regions, predict nonlinear accumulation of mutations in pseudogenes over time, and provide a possible explanation for the observed higher diversity of single nucleotide polymorphisms (SNPs) in the synonymous sites of exons compared to the noncoding regions.
Figures

Similar articles
-
Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: evidence for functional effects of synonymous polymorphisms.BMC Genomics. 2006 Aug 17;7:213. doi: 10.1186/1471-2164-7-213. BMC Genomics. 2006. PMID: 16916449 Free PMC article.
-
Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms.Genomics. 2006 Nov;88(5):527-34. doi: 10.1016/j.ygeno.2006.06.003. Epub 2006 Jul 24. Genomics. 2006. PMID: 16860534
-
Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes.Genome Res. 2010 Apr;20(4):447-57. doi: 10.1101/gr.098947.109. Epub 2010 Jan 26. Genome Res. 2010. PMID: 20103589 Free PMC article.
-
Functions of noncoding sequences in mammalian genomes.Biochemistry (Mosc). 2014 Dec;79(13):1442-69. doi: 10.1134/S0006297914130021. Biochemistry (Mosc). 2014. PMID: 25749159 Review.
-
Realizing the significance of noncoding functionality in clinical genomics.Exp Mol Med. 2018 Aug 7;50(8):1-8. doi: 10.1038/s12276-018-0087-0. Exp Mol Med. 2018. PMID: 30089779 Free PMC article. Review.
Cited by
-
Transcription-related mutations and GC content drive variation in nucleotide substitution rates across the genomes of Arabidopsis thaliana and Arabidopsis lyrata.BMC Evol Biol. 2007 Apr 23;7:66. doi: 10.1186/1471-2148-7-66. BMC Evol Biol. 2007. PMID: 17451608 Free PMC article.
-
Patterns in interspecies similarity correlate with nucleotide composition in mammalian 3'UTRs.Nucleic Acids Res. 2003 Sep 15;31(18):5433-9. doi: 10.1093/nar/gkg751. Nucleic Acids Res. 2003. PMID: 12954780 Free PMC article.
-
Signatures of selection and gene conversion associated with human color vision variation.Am J Hum Genet. 2004 Sep;75(3):363-75. doi: 10.1086/423287. Epub 2004 Jul 13. Am J Hum Genet. 2004. PMID: 15252758 Free PMC article.
-
A Novel Terminal-Repeat Retrotransposon in Miniature (TRIM) Is Massively Expressed in Echinococcus multilocularis Stem Cells.Genome Biol Evol. 2015 Jul 1;7(8):2136-53. doi: 10.1093/gbe/evv126. Genome Biol Evol. 2015. PMID: 26133390 Free PMC article.
-
Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects.J Mol Evol. 2005 Jun;60(6):748-63. doi: 10.1007/s00239-004-0222-5. Epub 2005 May 31. J Mol Evol. 2005. PMID: 15959677
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
