Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein
- PMID: 12730166
- PMCID: PMC154060
- DOI: 10.1128/JB.185.10.3068-3075.2003
Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein
Abstract
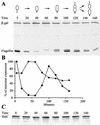
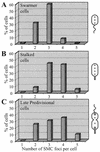
Structural maintenance of chromosomes proteins (SMCs) bind to DNA and function to ensure proper chromosome organization in both eukaryotes and bacteria. Caulobacter crescentus possesses a single SMC homolog that plays a role in organizing and segregating daughter chromosomes. Approximately 1,500 to 2,000 SMC molecules are present per cell during active growth, corresponding to one SMC complex per 6,000 to 8,000 bp of chromosomal DNA. Although transcription from the smc promoter is induced during early S phase, a cell cycle transcription pattern previously observed with multiple DNA replication and repair genes, the SMC protein is present throughout the entire cell cycle. Examination of the intracellular location of SMC showed that in swarmer cells, which do not replicate DNA, the protein forms two or three foci. Stalked cells, which are actively engaged in DNA replication, have three or four SMC foci per cell. The SMC foci appear randomly distributed in the cell. Many predivisional cells have bright polar SMC foci, which are lost upon cell division. Thus, chromosome compaction likely involves dynamic aggregates of SMC bound to DNA. The aggregation pattern changes as a function of the cell cycle both during and upon completion of chromosome replication.
Figures



References
-
- Akhmedov, A. T., C. Frei, M. Tsai-Pflugfelder, B. Kemper, S. M. Gasser, and R. Jessberger. 1998. Structural maintenance of chromosomes: protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem. 273:24088-24094. - PubMed
-
- Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. - PubMed
-
- Cobbe, N., and M. M. Heck. 2000. Review: SMCs in the world of chromosome biology—from prokaryotes to higher eukaryotes. J. Struct. Biol. 129:123-143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

