Repair system for noncanonical purines in Escherichia coli
- PMID: 12730170
- PMCID: PMC154070
- DOI: 10.1128/JB.185.10.3101-3110.2003
Repair system for noncanonical purines in Escherichia coli
Abstract
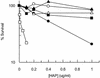
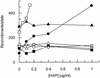
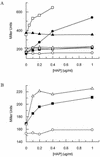
Exposure of Escherichia coli strains deficient in molybdopterin biosynthesis (moa) to the purine base N-6-hydroxylaminopurine (HAP) is mutagenic and toxic. We show that moa mutants exposed to HAP also exhibit elevated mutagenesis, a hyperrecombination phenotype, and increased SOS induction. The E. coli rdgB gene encodes a protein homologous to a deoxyribonucleotide triphosphate pyrophosphatase from Methanococcus jannaschii that shows a preference for purine base analogs. moa rdgB mutants are extremely sensitive to killing by HAP and exhibit increased mutagenesis, recombination, and SOS induction upon HAP exposure. Disruption of the endonuclease V gene, nfi, rescues the HAP sensitivity displayed by moa and moa rdgB mutants and reduces the level of recombination and SOS induction, but it increases the level of mutagenesis. Our results suggest that endonuclease V incision of DNA containing HAP leads to increased recombination and SOS induction and even cell death. Double-strand break repair mutants display an increase in HAP sensitivity, which can be reversed by an nfi mutation. This suggests that cell killing may result from an increase in double-strand breaks generated when replication forks encounter endonuclease V-nicked DNA. We propose a pathway for the removal of HAP from purine pools, from deoxynucleotide triphosphate pools, and from DNA, and we suggest a general model for excluding purine base analogs from DNA. The system for HAP removal consists of a molybdoenzyme, thought to detoxify HAP, a deoxyribonucleotide triphosphate pyrophosphatase that removes noncanonical deoxyribonucleotide triphosphates from replication precursor pools, and an endonuclease that initiates the removal of HAP from DNA.
Figures



References
-
- Arnold, D. A., and S. C. Kowalczykowski. 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 275:12261-12265. - PubMed
-
- Bhatnagar, S. K., L. C. Bullions, and M. J. Bessman. 1991. Characterization of the mutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266:9050-9054. - PubMed
-
- Clement, B., and T. Kunze. 1992. The reduction of 6-N-hydroxylaminopurine to adenine by xanthine oxidase. Biochem. Pharmacol. 44:1501-1509. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

