Genomewide view of gene silencing by small interfering RNAs
- PMID: 12730368
- PMCID: PMC164448
- DOI: 10.1073/pnas.1037853100
Genomewide view of gene silencing by small interfering RNAs
Abstract
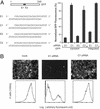
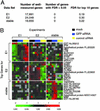
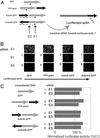
RNA interference (RNAi) is an evolutionarily conserved mechanism in plant and animal cells that directs the degradation of messenger RNAs homologous to short double-stranded RNAs termed small interfering RNA (siRNA). The ability of siRNA to direct gene silencing in mammalian cells has raised the possibility that siRNA might be used to investigate gene function in a high throughput fashion or to modulate gene expression in human diseases. The specificity of siRNA-mediated silencing, a critical consideration in these applications, has not been addressed on a genomewide scale. Here we show that siRNA-induced gene silencing of transient or stably expressed mRNA is highly gene-specific and does not produce secondary effects detectable by genomewide expression profiling. A test for transitive RNAi, extension of the RNAi effect to sequences 5' of the target region that has been observed in Caenorhabditis elegans, was unable to detect this phenomenon in human cells.
Figures
Comment in
-
The specifics of small interfering RNA specificity.Proc Natl Acad Sci U S A. 2003 May 27;100(11):6289-91. doi: 10.1073/pnas.1232238100. Epub 2003 May 16. Proc Natl Acad Sci U S A. 2003. PMID: 12754379 Free PMC article. No abstract available.
References
-
- Hannon, G. J. (2002) Nature 418, 244–251. - PubMed
-
- Zamore, P. D. (2002) Science 296, 1265–1269. - PubMed
-
- Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. - PubMed
-
- Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325–330. - PubMed
-
- Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

