Digital genotyping and haplotyping with polymerase colonies
- PMID: 12730373
- PMCID: PMC156303
- DOI: 10.1073/pnas.0936399100
Digital genotyping and haplotyping with polymerase colonies
Abstract
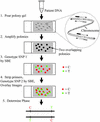
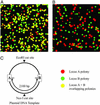
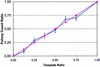
Polymerase colony (polony) technology amplifies multiple individual DNA molecules within a thin acrylamide gel attached to a microscope slide. Each DNA molecule included in the reaction produces an immobilized colony of double-stranded DNA. We genotype these polonies by performing single base extensions with dye-labeled nucleotides, and we demonstrate the accurate quantitation of two allelic variants. We also show that polony technology can determine the phase, or haplotype, of two single- nucleotide polymorphisms (SNPs) by coamplifying distally located targets on a single chromosomal fragment. We correctly determine the genotype and phase of three different pairs of SNPs. In one case, the distance between the two SNPs is 45 kb, the largest distance achieved to date without separating the chromosomes by cloning or somatic cell fusion. The results indicate that polony genotyping and haplotyping may play an important role in understanding the structure of genetic variation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

