The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress
- PMID: 12732516
- PMCID: PMC154493
- DOI: 10.1128/AEM.69.5.2512-2520.2003
The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress
Abstract
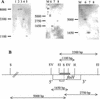
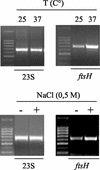
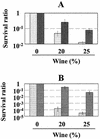
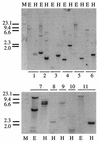
The wine bacterium Oenococcus oeni has to cope with harsh environmental conditions, including an acidic pH, a high alcoholic content, nonoptimal growth temperatures, and growth-inhibitory compounds such as fatty acids, phenolic acids, and tannins. We describe the characterization and cloning of the O. oeni ftsH gene, encoding a protease belonging to the ATP binding cassette protein superfamily. The O. oeni FtsH protein is closest in sequence similarity to the FtsH homologue of Lactococcus lactis. The O. oeni ftsH gene proved to be stress-responsive, since its expression increased at high temperatures or under osmotic shock. O. oeni FtsH protein function was tested in an Escherichia coli ftsH mutant strain, and consistent with the O. oeni ftsH gene expression pattern, the O. oeni FtsH protein provided protection for the E. coli ftsH mutant against heat shock. O. oeni and Bradyrhizobium japonicum FtsH proteins also triggered E. coli resistance to wine toxicity. Genes homologous to O. oeni ftsH were detected in many other lactic acid bacteria found in wine, suggesting that this type of gene constitutes a well-conserved stress-protective molecular device.
Figures




References
-
- Akiyama, Y., T. Ogura, and K. Ito. 1994. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention efficiency of a cytoplasmic reporter. J. Biol. Chem. 269:5218-5224. - PubMed
-
- Confalonieri, F., and M. Duguet. 1995. A 200-amino acid ATPase module in search of a basic function. Bioessays 17:639-650. - PubMed
-
- Edwards, C. G., R. B. Beelman, C. E. Bartley, and A. McConnell. 1990. Production of decanoic acid and other volatile compounds and the growth of yeast and malolactic bacteria during vinification. Am. J. Enol. Vitic. 41:48-56.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

