Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure
- PMID: 12732521
- PMCID: PMC154551
- DOI: 10.1128/AEM.69.5.2555-2562.2003
Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure
Abstract
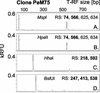
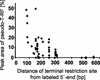
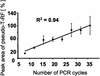
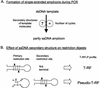
Terminal restriction fragment length polymorphism (T-RFLP) analysis of PCR-amplified genes is a widely used fingerprinting technique in molecular microbial ecology. In this study, we show that besides expected terminal restriction fragments (T-RFs), additional secondary T-RFs occur in T-RFLP analysis of amplicons from cloned 16S rRNA genes at high frequency. A total of 50% of 109 bacterial and 78% of 68 archaeal clones from the guts of cetoniid beetle larvae, using MspI and AluI as restriction enzymes, respectively, were affected by the presence of these additional T-RFs. These peaks were called "pseudo-T-RFs" since they can be detected as terminal fluorescently labeled fragments in T-RFLP analysis but do not represent the primary terminal restriction site as indicated by sequence data analysis. Pseudo-T-RFs were also identified in T-RFLP profiles of pure culture and environmental DNA extracts. Digestion of amplicons with the single-strand-specific mung bean nuclease prior to T-RFLP analysis completely eliminated pseudo-T-RFs. This clearly indicates that single-stranded amplicons are the reason for the formation of pseudo-T-RFs, most probably because single-stranded restriction sites cannot be cleaved by restriction enzymes. The strong dependence of pseudo-T-RF formation on the number of cycles used in PCR indicates that (partly) single-stranded amplicons can be formed during amplification of 16S rRNA genes. In a model, we explain how transiently formed secondary structures of single-stranded amplicons may render single-stranded amplicons accessible to restriction enzymes. The occurrence of pseudo-T-RFs has consequences for the interpretation of T-RFLP profiles from environmental samples, since pseudo-T-RFs may lead to an overestimation of microbial diversity. Therefore, it is advisable to establish 16S rRNA gene sequence clone libraries in parallel with T-RFLP analysis from the same sample and to check clones for their in vitro digestion T-RF pattern to facilitate the detection of pseudo-T-RFs.
Figures



Similar articles
-
Post-amplification Klenow fragment treatment alleviates PCR bias caused by partially single-stranded amplicons.J Microbiol Methods. 2005 Apr;61(1):69-75. doi: 10.1016/j.mimet.2004.11.002. J Microbiol Methods. 2005. PMID: 15676197
-
Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes.Appl Environ Microbiol. 2001 Apr;67(4):1893-901. doi: 10.1128/AEM.67.4.1893-1901.2001. Appl Environ Microbiol. 2001. PMID: 11282647 Free PMC article.
-
Separation of fluorescence-labelled terminal restriction fragment DNA on a two-dimensional gel (T-RFs-2D) - an efficient approach for microbial consortium characterization.Environ Microbiol. 2011 Sep;13(9):2565-75. doi: 10.1111/j.1462-2920.2011.02527.x. Epub 2011 Aug 8. Environ Microbiol. 2011. PMID: 21824243
-
Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities.Appl Microbiol Biotechnol. 2008 Sep;80(3):365-80. doi: 10.1007/s00253-008-1565-4. Epub 2008 Jul 22. Appl Microbiol Biotechnol. 2008. PMID: 18648804 Review.
-
Technicalities and Glitches of Terminal Restriction Fragment Length Polymorphism (T-RFLP).Indian J Microbiol. 2014 Sep;54(3):255-61. doi: 10.1007/s12088-014-0461-0. Epub 2014 Mar 9. Indian J Microbiol. 2014. PMID: 24891731 Free PMC article. Review.
Cited by
-
Evaluating the assignment of alkB terminal restriction fragments and sequence types to distinct bacterial taxa.Appl Environ Microbiol. 2013 May;79(9):3129-32. doi: 10.1128/AEM.04028-12. Epub 2013 Mar 1. Appl Environ Microbiol. 2013. PMID: 23455350 Free PMC article.
-
Monthly to interannual variability of microbial eukaryote assemblages at four depths in the eastern North Pacific.ISME J. 2014 Mar;8(3):515-530. doi: 10.1038/ismej.2013.173. Epub 2013 Oct 31. ISME J. 2014. PMID: 24173457 Free PMC article.
-
Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones.Appl Environ Microbiol. 2004 Mar;70(3):1787-94. doi: 10.1128/AEM.70.3.1787-1794.2004. Appl Environ Microbiol. 2004. PMID: 15006805 Free PMC article.
-
Spatial Variation of the Gut Microbiota in Broiler Chickens as Affected by Dietary Available Phosphorus and Assessed by T-RFLP Analysis and 454 Pyrosequencing.PLoS One. 2015 Nov 20;10(11):e0143442. doi: 10.1371/journal.pone.0143442. eCollection 2015. PLoS One. 2015. PMID: 26588075 Free PMC article.
-
Humic acids buffer the effects of urea on soil ammonia oxidizers and potential nitrification.Soil Biol Biochem. 2009 Aug;41(8):1612-1621. doi: 10.1016/j.soilbio.2009.04.023. Soil Biol Biochem. 2009. PMID: 22267875 Free PMC article.
References
-
- Clement, B. G., L. E. Kehl, K. L. Debord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

