Cloning and sequencing of the histidine decarboxylase genes of gram-negative, histamine-producing bacteria and their application in detection and identification of these organisms in fish
- PMID: 12732523
- PMCID: PMC154508
- DOI: 10.1128/AEM.69.5.2568-2579.2003
Cloning and sequencing of the histidine decarboxylase genes of gram-negative, histamine-producing bacteria and their application in detection and identification of these organisms in fish
Abstract
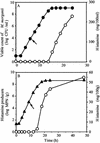
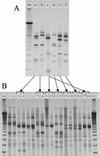
The use of molecular tools for early and rapid detection of gram-negative histamine-producing bacteria is important for preventing the accumulation of histamine in fish products. To date, no molecular detection or identification system for gram-negative histamine-producing bacteria has been developed. A molecular method that allows the rapid detection of gram-negative histamine producers by PCR and simultaneous differentiation by single-strand conformation polymorphism (SSCP) analysis using the amplification product of the histidine decarboxylase genes (hdc) was developed. A collection of 37 strains of histamine-producing bacteria (8 reference strains from culture collections and 29 isolates from fish) and 470 strains of non-histamine-producing bacteria isolated from fish were tested. Histamine production of bacteria was determined by paper chromatography and confirmed by high-performance liquid chromatography. Among 37 strains of histamine-producing bacteria, all histidine-decarboxylating gram-negative bacteria produced a PCR product, except for a strain of Citrobacter braakii. In contrast, none of the non-histamine-producing strains (470 strains) produced an amplification product. Specificity of the amplification was further confirmed by sequencing the 0.7-kbp amplification product. A phylogenetic tree of the isolates constructed using newly determined sequences of partial hdc was similar to the phylogenetic tree generated from 16S ribosomal DNA sequences. Histamine accumulation occurred when PCR amplification of hdc was positive in all of fish samples tested and the presence of powerful histamine producers was confirmed by subsequent SSCP identification. The potential application of the PCR-SSCP method as a rapid monitoring tool is discussed.
Figures


References
-
- Akkermans, A. D. L., M. S. Mirza, H. J. M. Harmsen, H. J. Blok, P. R. Heron, A. Sessitsch, and W. M. Akkerlnans. 1994. Molecular ecology of microbes: review of promises, pitfalls, and true progress. FEMS Microbiol. Rev: 15:185-194.
-
- Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. - PubMed
-
- Ben-Gigirey, B., J. M. V. B. de Sousa, T. G. Villa, and J. Barros-Velazquez. 1998. Changes in biogenic amines and microbiological analysis in albacore (Thunnus alalunga) muscle during frozen storage. J. Food Prot. 61:608-615. - PubMed
-
- Bover-Cid, S., and W. H. Holzapfel. 1999. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53:33-41. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

