Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality?
- PMID: 12732530
- PMCID: PMC154555
- DOI: 10.1128/AEM.69.5.2624-2630.2003
Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality?
Abstract
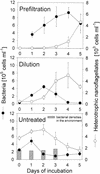
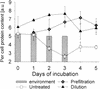
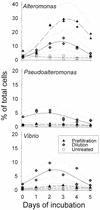
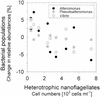
We studied the growth of six culturable bacterial lineages from coastal North Sea picoplankton in environmental samples under different incubation conditions. The grazing pressure of heterotrophic nanoflagellates (HNF) was reduced either by double prefiltration through 0.8- micro m-pore-size filters or by 10-fold dilutions with 0.2- micro m (pore-size) prefiltered seawater. We hypothesized that those gamma-proteobacterial genera that are rapidly enriched would also be most strongly affected by HNF regrowth. In the absence of HNF, the mean protein content per bacterial cell increased in both treatments compared to environmental samples, whereas the opposite trend was found in incubations of unaltered seawater. Significant responses to the experimental manipulations were observed in Alteromonas, Pseudoalteromonas, and Vibrio populations. No treatment-specific effects could be detected for members of the Roseobacter group, the Cytophaga latercula-C. marinoflava lineage, or the NOR5 clade. Statistical analysis confirmed a transient increase in the proportions of Alteromonas, Pseudoalteromonas, and Vibrio cells at reduced HNF densities only, followed by an overproportional decline during the phase of HNF regrowth. Cells from these genera were significantly larger than the community average in the dilution treatments, and changes in their relative abundances were negatively correlated with HNF densities. Our findings suggest that bacteria affiliated with frequently isolated genera such as Alteromonas, Pseudoalteromonas, and Vibrio might be rare in coastal North Sea picoplankton because their rapid growth response to changing environmental conditions is counterbalanced by a higher grazing mortality.
Figures


References
-
- Button, D. K., B. R. Robertson, P. W. Lepp, and T. M. Schmidt. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467-4476. - PMC - PubMed
-
- Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. - PubMed
-
- Del Giorgio, P. A., J. M. Gasol, D. Vaque, P. Mura, S. Agusti, and C. M. Duarte. 1996. Bacterioplankton community structure: protists control net production and the propoportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41:1169-1179.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

