Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments
- PMID: 12732534
- PMCID: PMC154554
- DOI: 10.1128/AEM.69.5.2657-2663.2003
Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments
Abstract
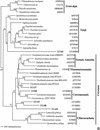
Microeukaryotes in oxygen-depleted environments are among the most diverse, as well as the least studied, organisms. We conducted a cultivation-independent, small-subunit (SSU) rRNA-based survey of microeukaryotes in suboxic waters and anoxic sediments in the great Sippewisset salt marsh, Cape Cod, Mass. We generated two clone libraries and analyzed approximately 300 clones, which contained a large diversity of microeukaryotic SSU rRNA signatures. Only a few of these signatures were closely related (sequence similarity of >97%) to the sequences reported earlier. The bulk of our sequences represented deep novel branches within green algae, fungi, cercozoa, stramenopiles, alveolates, euglenozoa and unclassified flagellates. In addition, a significant number of detected rRNA sequences exhibited no affiliation to known organisms and sequences and thus represent novel lineages of the highest taxonomical order, most of them branching off the base of the global phylogenetic tree. This suggests that oxygen-depleted environments harbor diverse communities of novel organisms, which may provide an interesting window into the early evolution of eukaryotes.
Figures




References
-
- Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [Correction, 3:15.]. - PMC - PubMed
-
- Colwell, R. R., and D. J. Grimes. 2000. Semantics and strategies. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment, ASM Press, Washington D.C.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

