Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster
- PMID: 12732539
- PMCID: PMC154558
- DOI: 10.1128/AEM.69.5.2699-2706.2003
Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster
Abstract
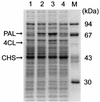
In plants, chalcones are precursors for a large number of flavonoid-derived plant natural products and are converted to flavanones by chalcone isomerase or nonenzymatically. Chalcones are synthesized from tyrosine and phenylalanine via the phenylpropanoid pathway involving phenylalanine ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate:coenzyme A ligase (4CL), and chalcone synthase (CHS). For the purpose of production of flavanones in Escherichia coli, three sets of an artificial gene cluster which contained three genes of heterologous origins--PAL from the yeast Rhodotorula rubra, 4CL from the actinomycete Streptomyces coelicolor A3(2), and CHS from the licorice plant Glycyrrhiza echinata--were constructed. The constructions of the three sets were done as follows: (i) PAL, 4CL, and CHS were placed in that order under the control of the T7 promoter (P(T7)) and the ribosome-binding sequence (RBS) in the pET vector, where the initiation codons of 4CL and CHS were overlapped with the termination codons of the preceding genes; (ii) the three genes were transcribed by a single P(T7) in front of PAL, and each of the three contained the RBS at appropriate positions; and (iii) all three genes contained both P(T7) and the RBS. These pathways bypassed C4H, a cytochrome P-450 hydroxylase, because the bacterial 4CL enzyme ligated coenzyme A to both cinnamic acid and 4-coumaric acid. E. coli cells containing the gene clusters produced two flavanones, pinocembrin from phenylalanine and naringenin from tyrosine, in addition to their precursors, cinnamic acid and 4-coumaric acid. Of the three sets, the third gene cluster conferred on the host the highest ability to produce the flavanones. This is a new metabolic engineering technique for the production in bacteria of a variety of compounds of plant and animal origin.
Figures




References
-
- Baedeker, M., and G. E. Schulz. 1999. Overexpression of a designed 2.2 kb gene of eukaryotic phenylalanine ammonia-lyase in E. coli. FEBS Lett. 457:57-60. - PubMed
-
- Davis, M. S., J. Solbiati, and J. E. Cronan, Jr. 2000. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 275:28593-28598. - PubMed
-
- Dixon, R. A., and C. L. Steele. 1999. Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends Plant Sci. 4:394-400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

