In situ accessibility of Saccharomyces cerevisiae 26S rRNA to Cy3-labeled oligonucleotide probes comprising the D1 and D2 domains
- PMID: 12732564
- PMCID: PMC154520
- DOI: 10.1128/AEM.69.5.2899-2905.2003
In situ accessibility of Saccharomyces cerevisiae 26S rRNA to Cy3-labeled oligonucleotide probes comprising the D1 and D2 domains
Abstract
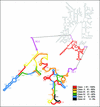
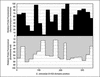
Fluorescence in situ hybridization (FISH) has proven to be most useful for the identification of microorganisms. However, species-specific oligonucleotide probes often fail to give satisfactory results. Among the causes leading to low hybridization signals is the reduced accessibility of the targeted rRNA site to the oligonucleotide, mainly for structural reasons. In this study we used flow cytometry to determine whole-cell fluorescence intensities with a set of 32 Cy3-labeled oligonucleotide probes covering the full length of the D1 and D2 domains in the 26S rRNA of Saccharomyces cerevisiae PYCC 4455(T). The brightest signal was obtained with a probe complementary to positions 223 to 240. Almost half of the probes conferred a fluorescence intensity above 60% of the maximum, whereas only one probe could hardly detect the cells. The accessibility map based on the results obtained can be extrapolated to other yeasts, as shown experimentally with 27 additional species (14 ascomycetes and 13 basidiomycetes). This work contributes to a more rational design of species-specific probes for yeast identification and monitoring.
Figures


References
-
- Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. - PubMed
-
- Crockett, A. O., and C. T. Wittwer. 2001. Fluorescein-labeled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem. 290:89-97. - PubMed
-
- Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. E vol. Microbiol. 50:1351-1371. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

