Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis
- PMID: 12732614
- PMCID: PMC2172928
- DOI: 10.1083/jcb.200302072
Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis
Abstract
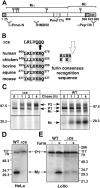
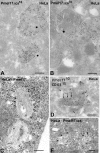
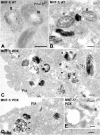
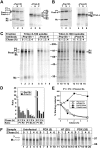
Lysosome-related organelles are cell type-specific intracellular compartments with distinct morphologies and functions. The molecular mechanisms governing the formation of their unique structural features are not known. Melanosomes and their precursors are lysosome-related organelles that are characterized morphologically by intralumenal fibrous striations upon which melanins are polymerized. The integral membrane protein Pmel17 is a component of the fibrils and can nucleate their formation in the absence of other pigment cell-specific proteins. Here, we show that formation of intralumenal fibrils requires cleavage of Pmel17 by a furin-like proprotein convertase (PC). As in the generation of amyloid, proper cleavage of Pmel17 liberates a lumenal domain fragment that becomes incorporated into the fibrils; longer Pmel17 fragments generated in the absence of PC activity are unable to form organized fibrils. Our results demonstrate that PC-dependent cleavage regulates melanosome biogenesis by controlling the fibrillogenic activity of a resident protein. Like the pathologic process of amyloidogenesis, the formation of other tissue-specific organelle structures may be similarly dependent on proteolytic activation of physiological fibrillogenic substrates.
Figures


Comment in
-
Amyloid as a natural product.J Cell Biol. 2003 May 12;161(3):461-2. doi: 10.1083/jcb.200304074. J Cell Biol. 2003. PMID: 12743097 Free PMC article. Review.
References
-
- Adema, G.J., A.J. de Boer, A.M. Vogel, W.A.M. Loenen, and C.G. Figdor. 1994. Molecular characterization of the melanocyte lineage-specific antigen gp100. J. Biol. Chem. 269:20126–20133. - PubMed
-
- Anderson, E.D., L. Thomas, J.S. Hayflick, and G. Thomas. 1993. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed α1-antitrypsin variant. J. Biol. Chem. 268:24887–24891. - PubMed
-
- Badman, M.K., K.I. Shennan, J.L. Jermany, K. Docherty, and A. Clark. 1996. Processing of pro-islet amyloid polypeptide (proIAPP) by the prohormone convertase PC2. FEBS Lett. 378:227–231. - PubMed
-
- Berson, J.F., D.W. Frank, P.A. Calvo, B.M. Bieler, and M.S. Marks. 2000. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J. Biol. Chem. 275:12281–12289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

