Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways
- PMID: 12732713
- PMCID: PMC164498
- DOI: 10.1073/pnas.1131932100
Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways
Abstract
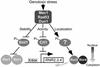
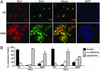
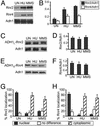
The fidelity of DNA replication and repair processes is critical for maintenance of genomic stability. Ribonucleotide reductase (RNR) catalyzes the rate-limiting step in dNTP production and thus plays an essential role in DNA synthesis. The level and activity of RNR are highly regulated by the cell cycle and DNA damage checkpoints, which maintain optimal dNTP pools required for genetic fidelity. RNRs are composed of a large subunit that binds the nucleoside diphosphate substrates and allosteric effectors and a small subunit that houses the di-iron tyrosyl radical cofactor essential for the reduction process. In Saccharomyces cerevisiae, there are two large subunits (Rnr1 and Rnr3) and two small subunits (Rnr2 and Rnr4). Here we report the subcellular localization of Rnr1-4 during normal cell growth and the redistribution of Rnr2 and Rnr4 in response to DNA damage and replicational stress. During the normal cell cycle, Rnr1 and Rnr3 are predominantly localized to the cytoplasm and Rnr2 and Rnr4 are predominantly present in the nucleus. Under genotoxic stress, Rnr2 and Rnr4 become redistributed to the cytoplasm in a checkpoint-dependent manner. Subcellular redistribution of Rnr2 and Rnr4 can occur in the absence of the transcriptional induction of the RNR genes after DNA damage and likely represents a posttranslational event. These results suggest a mechanism by which DNA damage checkpoint modulates RNR activity through the temporal and spatial regulation of its subunits.
Figures



References
-
- Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. - PubMed
-
- Khanna, K. K. & Jackson, S. P. (2001) Nat. Genet. 27, 247–254. - PubMed
-
- Kolodner, R. D., Putnam, C. D. & Myung, K. (2002) Science 297, 552–577. - PubMed
-
- Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643–649. - PubMed
-
- Abraham, R. T. (2001) Genes Dev. 15, 2177–2196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

