Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7
- PMID: 12734215
- PMCID: PMC154664
- DOI: 10.1128/JCM.41.5.1843-1849.2003
Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7
Abstract
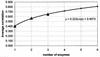
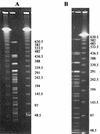
Pulsed-field gel electrophoresis (PFGE) has been used extensively to investigate the epidemiology of Escherichia coli O157:H7, although it has not been evaluated as a tool for establishing genetic relationships. This is a critical issue when molecular genetic data are used to make inferences about pathogen dissemination. To evaluate this further, genomic DNAs from 62 isolates of E. coli O157:H7 from different cattle herds were digested with XbaI and BlnI and subjected to PFGE. The correlation between the similarity coefficients for these two enzymes was only 0.53. Four additional restriction enzymes (NheI, PacI, SfiI, and SpeI) were used with DNAs from a subset of 14 isolates. The average correlations between similarity coefficients using sets of one, two, and three enzymes were 0.405, 0.568, and 0.648, respectively. Probing with lambda DNA demonstrated that some DNA fragments migrated equal distances in the gel but were composed of nonhomologous genetic material. Genome sequence data from EDL933 indicated that 40 PFGE fragments would be expected from complete XbaI digestion, yet only 19 distinguishable fragments were visible. Two reasons that similarity coefficients from single-enzyme PFGE are poor measures of relatedness (and hence are poorly correlated with other enzymes) are evident from this study: (i) matching bands do not always represent homologous genetic material and (ii) there are limitations to the power of PFGE to resolve bands of nearly identical size. The findings of the present study indicate that if genetic relationships must be inferred in the absence of epidemiologic data, six or more restriction enzymes would be needed to provide a reasonable estimate using PFGE.
Figures


Similar articles
-
Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting.Appl Environ Microbiol. 2008 Nov;74(21):6606-15. doi: 10.1128/AEM.00985-08. Epub 2008 Sep 12. Appl Environ Microbiol. 2008. PMID: 18791027 Free PMC article.
-
Comparison of the molecular genotypes of Escherichia coli O157:H7 from the hides of beef cattle in different regions of North America.J Food Prot. 2007 Jul;70(7):1622-6. doi: 10.4315/0362-028x-70.7.1622. J Food Prot. 2007. PMID: 17685334
-
Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease.J Clin Microbiol. 2002 Aug;40(8):2806-12. doi: 10.1128/JCM.40.8.2806-2812.2002. J Clin Microbiol. 2002. PMID: 12149334 Free PMC article.
-
Polymorphic amplified typing sequences provide a novel approach to Escherichia coli O157:H7 strain typing.J Clin Microbiol. 2002 Apr;40(4):1152-9. doi: 10.1128/JCM.40.4.1152-1159.2002. J Clin Microbiol. 2002. PMID: 11923324 Free PMC article.
-
The Application of Pulsed Field Gel Electrophoresis in Clinical Studies.J Clin Diagn Res. 2016 Jan;10(1):DE01-4. doi: 10.7860/JCDR/2016/15718.7043. Epub 2016 Jan 1. J Clin Diagn Res. 2016. PMID: 26894068 Free PMC article. Review.
Cited by
-
Comparison of subtyping methods for differentiating Salmonella enterica serovar Typhimurium isolates obtained from food animal sources.J Clin Microbiol. 2006 Oct;44(10):3569-77. doi: 10.1128/JCM.00745-06. J Clin Microbiol. 2006. PMID: 17021084 Free PMC article.
-
Escherichia coil cluster evaluation.Emerg Infect Dis. 2007 Mar;13(3):520. doi: 10.3201/eid1303.041085. Emerg Infect Dis. 2007. PMID: 17552125 Free PMC article. No abstract available.
-
Comparison of rectoanal mucosal swab cultures and fecal cultures for determining prevalence of Escherichia coli O157:H7 in feedlot cattle.Appl Environ Microbiol. 2005 Oct;71(10):6431-3. doi: 10.1128/AEM.71.10.6431-6433.2005. Appl Environ Microbiol. 2005. PMID: 16204574 Free PMC article.
-
CTX-M-3 and CTX-M-15 extended-spectrum beta-lactamases in isolates of Escherichia coli from a hospital in Algiers, Algeria.J Clin Microbiol. 2006 Dec;44(12):4584-6. doi: 10.1128/JCM.01445-06. Epub 2006 Sep 20. J Clin Microbiol. 2006. PMID: 16988017 Free PMC article.
-
Whole Genome Sequencing for Genomics-Guided Investigations of Escherichia coli O157:H7 Outbreaks.Front Microbiol. 2016 Jun 30;7:985. doi: 10.3389/fmicb.2016.00985. eCollection 2016. Front Microbiol. 2016. PMID: 27446025 Free PMC article.
References
-
- Amavisit, P., P. F. Markham, D. Lightfoot, K. G. Whithear, and G. F. Browning. 2001. Molecular epidemiology of Salmonella Heidelberg in an equine hospital. Vet. Microbiol. 80:85-98. - PubMed
-
- Barkocy-Gallagher, G. A., T. M. Arthur, G. R. Siragusa, J. E. Keen, R. O. Elder, W. W. Laegreid, and M. Koohmaraie. 2001. Genotypic analysis of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the Midwestern states of the United States. Appl. Environ. Microbiol. 67:3810-3818. - PMC - PubMed
-
- Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. - PMC - PubMed
-
- Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, R. Baron, and J. Kobayashi. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers: the Washington experience. JAMA 272:1349-1353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

