Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays
- PMID: 12734257
- PMCID: PMC154688
- DOI: 10.1128/JCM.41.5.2113-2125.2003
Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays
Abstract
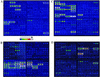
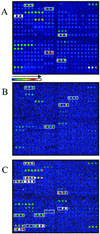
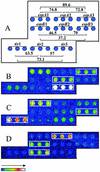
One approach to the accurate determination of the pathogenic potential (pathotype) of isolated Escherichia coli strains would be through a complete assessment of each strain for the presence of all known E. coli virulence factors. To accomplish this, an E. coli virulence factor DNA microarray composed of 105 DNA PCR amplicons printed on glass slides and arranged in eight subarrays corresponding to different E. coli pathotypes was developed. Fluorescently labeled genomic DNAs from E. coli strains representing known pathotypes were initially hybridized to the virulence gene microarrays for both chip optimization and validation. Hybridization pattern analysis with clinical isolates permitted a rapid assessment of their virulence attributes and determination of the pathogenic group to which they belonged. Virulence factors belonging to two different pathotypes were detected in one human E. coli isolate (strain H87-5406). The microarray was also tested for its ability to distinguish among phylogenetic groups of genes by using gene probes derived from the attaching-and-effacing locus (espA, espB, tir). After hybridization with these probes, we were able to distinguish E. coli strains harboring espA, espB, and tir sequences closely related to the gene sequences of an enterohemorrhagic strain (EDL933), a human enteropathogenic strain (E2348/69), or an animal enteropathogenic strain (RDEC-1). Our results show that the virulence factor microarray is a powerful tool for diagnosis-based studies and that the concept is useful for both gene quantitation and subtyping. Additionally, the multitude of virulence genes present on the microarray should greatly facilitate the detection of virulence genes acquired by horizontal transfer and the identification of emerging pathotypes.
Figures


Similar articles
-
STEC-EPEC oligonucleotide microarray: a new tool for typing genetic variants of the LEE pathogenicity island of human and animal Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) strains.Clin Chem. 2006 Feb;52(2):192-201. doi: 10.1373/clinchem.2005.059766. Epub 2005 Dec 29. Clin Chem. 2006. PMID: 16384888
-
Differentiation of Escherichia coli pathotypes by oligonucleotide spotted array.J Clin Microbiol. 2006 Apr;44(4):1495-501. doi: 10.1128/JCM.44.4.1495-1501.2006. J Clin Microbiol. 2006. PMID: 16597882 Free PMC article.
-
Virulence typing of Escherichia coli using microarrays.Mol Cell Probes. 2002 Oct;16(5):371-8. doi: 10.1006/mcpr.2002.0437. Mol Cell Probes. 2002. PMID: 12477441
-
Characterization of two virulence proteins secreted by rabbit enteropathogenic Escherichia coli, EspA and EspB, whose maximal expression is sensitive to host body temperature.Infect Immun. 1997 Sep;65(9):3547-55. doi: 10.1128/iai.65.9.3547-3555.1997. Infect Immun. 1997. PMID: 9284118 Free PMC article.
-
Are Escherichia coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing?Front Cell Infect Microbiol. 2016 Nov 18;6:141. doi: 10.3389/fcimb.2016.00141. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27917373 Free PMC article. Review.
Cited by
-
Phenotypic and Genotypic Characterization of Escherichia coli Isolated from Untreated Surface Waters.Open Microbiol J. 2013;7:9-19. doi: 10.2174/1874285801307010009. Epub 2013 Feb 22. Open Microbiol J. 2013. PMID: 23539437 Free PMC article.
-
A virulence and antimicrobial resistance DNA microarray detects a high frequency of virulence genes in Escherichia coli isolates from Great Lakes recreational waters.Appl Environ Microbiol. 2006 Jun;72(6):4200-6. doi: 10.1128/AEM.00137-06. Appl Environ Microbiol. 2006. PMID: 16751532 Free PMC article.
-
Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs.Infect Immun. 2006 Aug;74(8):4778-92. doi: 10.1128/IAI.00067-06. Infect Immun. 2006. PMID: 16861666 Free PMC article.
-
To kill or to be killed: pangenome analysis of Escherichia coli strains reveals a tailocin specific for pandemic ST131.BMC Biol. 2022 Jun 16;20(1):146. doi: 10.1186/s12915-022-01347-7. BMC Biol. 2022. PMID: 35710371 Free PMC article.
-
Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis.J Clin Microbiol. 2008 Jul;46(7):2280-90. doi: 10.1128/JCM.01752-07. Epub 2008 May 7. J Clin Microbiol. 2008. PMID: 18463209 Free PMC article.
References
-
- An, H., J. M. Fairbrother, C. Desautels, and J. Harel. 1999. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473:179-184. - PubMed
-
- Anderson, J. D., A. J. MacNab, W. R. Gransden, S. M. Damm, W. M. Johnson, and H. Lior. 1987. Gastroenteritis and encephalopathy associated with a strain of Escherichia coli O55:K59:H4 that produced a cytolethal distending toxin. Pediatr. Infect. Dis. J. 6:1135-1136. - PubMed
-
- Bach, S., A. de Almeida, and E. Carniel. 2000. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183:289-294. - PubMed
-
- Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

