Application of a Colorimetric Assay to Identify Putative Ribofuranosylaminobenzene 5'-Phosphate Synthase Genes Expressed with Activity in Escherichia coli
- PMID: 12734554
- PMCID: PMC152576
- DOI: 10.1251/bpo48
Application of a Colorimetric Assay to Identify Putative Ribofuranosylaminobenzene 5'-Phosphate Synthase Genes Expressed with Activity in Escherichia coli
Abstract
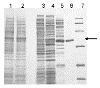
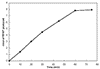
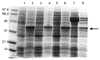
Tetrahydromethanopterin (H(4)MPT) is a tetrahydrofolate analog originally discovered in methanogenic archaea, but later found in other archaea and bacteria. The extent to which H(4)MPT occurs among living organisms is unknown. The key enzyme which distinguishes the biosynthetic pathways of H(4)MPT and tetrahydrofolate is ribofuranosylaminobenzene 5'-phosphate synthase (RFAP synthase). Given the importance of RFAP synthase in H(4)MPT biosynthesis, the identification of putative RFAP synthase genes and measurement of RFAP synthase activity would provide an indication of the presence of H(4)MPT in untested microorganisms. Investigation of putative archaeal RFAP synthase genes has been hampered by the tendency of the resulting proteins to form inactive inclusion bodies in Escherichia coli. The current work describes a colorimetric assay for measuring RFAP synthase activity, and two modified procedures for expressing recombinant RFAP synthase genes to produce soluble, active enzyme. By lowering the incubation temperature during expression, RFAP synthase from Archaeoglobus fulgidus was produced in E. coli and purified to homogeneity. The production of active RFAP synthase from Methanothermobacter thermautotrophicus was achieved by coexpression of the gene MTH0830 with a molecular chaperone. This is the first direct biochemical identification of a methanogen gene that codes for an active RFAP synthase.
Figures



Similar articles
-
Purification, kinetic characterization, and site-directed mutagenesis of Methanothermobacter thermautotrophicus RFAP Synthase Produced in Escherichia coli.AIMS Microbiol. 2019 Jul 23;5(3):186-204. doi: 10.3934/microbiol.2019.3.186. eCollection 2019. AIMS Microbiol. 2019. PMID: 31663056 Free PMC article.
-
Purification, overproduction, and partial characterization of beta-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway.J Bacteriol. 2002 Aug;184(16):4442-8. doi: 10.1128/JB.184.16.4442-4448.2002. J Bacteriol. 2002. PMID: 12142414 Free PMC article.
-
Bifunctional CTP:inositol-1-phosphate cytidylyltransferase/CDP-inositol:inositol-1-phosphate transferase, the key enzyme for di-myo-inositol-phosphate synthesis in several (hyper)thermophiles.J Bacteriol. 2007 Aug;189(15):5405-12. doi: 10.1128/JB.00465-07. Epub 2007 May 25. J Bacteriol. 2007. PMID: 17526717 Free PMC article.
-
Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism.Biochem J. 2000 Sep 15;350 Pt 3(Pt 3):609-29. Biochem J. 2000. PMID: 10970772 Free PMC article. Review.
-
Inositol-1-phosphate synthase from Archaeoglobus fulgidus is a class II aldolase.Biochemistry. 2000 Oct 10;39(40):12415-23. doi: 10.1021/bi001517q. Biochemistry. 2000. PMID: 11015222
Cited by
-
Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.Acta Crystallogr F Struct Biol Commun. 2014 Nov;70(Pt 11):1472-9. doi: 10.1107/S2053230X1402130X. Epub 2014 Oct 25. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25372812 Free PMC article.
-
Characterization of two methanopterin biosynthesis mutants of Methylobacterium extorquens AM1 by use of a tetrahydromethanopterin bioassay.J Bacteriol. 2004 Mar;186(5):1565-70. doi: 10.1128/JB.186.5.1565-1570.2004. J Bacteriol. 2004. PMID: 14973120 Free PMC article.
-
Discovery and characterization of the first archaeal dihydromethanopterin reductase, an iron-sulfur flavoprotein from Methanosarcina mazei.J Bacteriol. 2014 Jan;196(2):203-9. doi: 10.1128/JB.00457-13. Epub 2013 Aug 30. J Bacteriol. 2014. PMID: 23995635 Free PMC article.
-
Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance.Microbiol Mol Biol Rev. 2016 Dec 28;81(1):e00040-16. doi: 10.1128/MMBR.00040-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 28031352 Free PMC article. Review.
-
Purification, kinetic characterization, and site-directed mutagenesis of Methanothermobacter thermautotrophicus RFAP Synthase Produced in Escherichia coli.AIMS Microbiol. 2019 Jul 23;5(3):186-204. doi: 10.3934/microbiol.2019.3.186. eCollection 2019. AIMS Microbiol. 2019. PMID: 31663056 Free PMC article.
References
-
- Thauer RK, Hedderich R, Fischer R. Reactions and enzymes involved in methanogenesis from CO2 and H2. In: Ferry JG, editor. Methanogenesis. New York and London: Chapman Hall; 1993; p. 209-237.
-
- Van Beelen P, Labro JF, Keltjens JT, Geerts WM, Vogels GD, Laarhoven WM, Guijt W, Haasnoot CAG. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur J Biochem. 1984;139:359–365. - PubMed
-
- Möller-Zinkhan D, Börner G, Thauer RK. Function of methanofuran, tetrahydromethanopterin, and coenzyme F420 in Archaeoglobus fulgidus. Arch Microbiol. 1989;152:362–368.
LinkOut - more resources
Full Text Sources

