Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development
- PMID: 12736220
- PMCID: PMC3360973
- DOI: 10.1242/dev.00505
Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development
Abstract
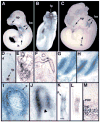
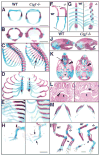
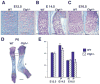
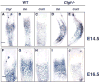
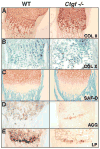
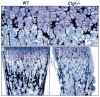
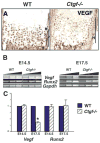
Coordinated production and remodeling of the extracellular matrix is essential during development. It is of particular importance for skeletogenesis, as the ability of cartilage and bone to provide structural support is determined by the composition and organization of the extracellular matrix. Connective tissue growth factor (CTGF, CCN2) is a secreted protein containing several domains that mediate interactions with growth factors, integrins and extracellular matrix components. A role for CTGF in extracellular matrix production is suggested by its ability to mediate collagen deposition during wound healing. CTGF also induces neovascularization in vitro, suggesting a role in angiogenesis in vivo. To test whether CTGF is required for extracellular matrix remodeling and/or angiogenesis during development, we examined the pattern of Ctgf expression and generated Ctgf-deficient mice. Ctgf is expressed in a variety of tissues in midgestation embryos, with highest levels in vascular tissues and maturing chondrocytes. We confirmed that CTGF is a crucial regulator of cartilage extracellular matrix remodeling by generating Ctgf(-/-) mice. Ctgf deficiency leads to skeletal dysmorphisms as a result of impaired chondrocyte proliferation and extracellular matrix composition within the hypertrophic zone. Decreased expression of specific extracellular matrix components and matrix metalloproteinases suggests that matrix remodeling within the hypertrophic zones in Ctgf mutants is defective. The mutant phenotype also revealed a role for Ctgf in growth plate angiogenesis. Hypertrophic zones of Ctgf mutant growth plates are expanded, and endochondral ossification is impaired. These defects are linked to decreased expression of vascular endothelial growth factor (VEGF) in the hypertrophic zones of Ctgf mutants. These results demonstrate that CTGF is important for cell proliferation and matrix remodeling during chondrogenesis, and is a key regulator coupling extracellular matrix remodeling to angiogenesis at the growth plate.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

