RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs
- PMID: 12736298
- PMCID: PMC156043
- DOI: 10.1093/nar/gkg361
RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs
Erratum in
- Nucleic Acids Res. 2013 Apr;41(8):4740
Abstract
By generating a specialised cDNA library from four different developmental stages of Drosophila melanogaster, we have identified 66 candidates for small non-messenger RNAs (snmRNAs) and have confirmed their expression by northern blot analysis. Thirteen of them were expressed at certain stages of D.melanogaster development, only. Thirty-five species belong to the class of small nucleolar RNAs (snoRNAs), divided into 15 members from the C/D subclass and 20 members from the H/ACA subclass, which mostly guide 2'-O-methylation and pseudouridylation, respectively, of rRNA and snRNAs. These also include two outstanding C/D snoRNAs, U3 and U14, both functioning as pre-rRNA chaperones. Surprisingly, the sequence of the Drosophila U14 snoRNA reflects a major change of function of this snoRNA in Diptera relative to yeast and vertebrates. Among the 22 snmRNAs lacking known sequence and structure motifs, five were located in intergenic regions, two in introns, five in untranslated regions of mRNAs, eight were derived from open reading frames, and two were transcribed opposite to an intron. Interestingly, detection of two RNA species from this group implies that certain snmRNA species are processed from alternatively spliced pre-mRNAs. Surprisingly, a few snmRNA sequences could not be found on the published D.melanogaster genome, which might suggest that more snmRNA genes (as well as mRNAs) are hidden in unsequenced regions of the genome.
Figures


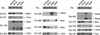




References
-
- Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

