Development and validation of a T7 based linear amplification for genomic DNA
- PMID: 12740028
- PMCID: PMC161798
- DOI: 10.1186/1471-2164-4-19
Development and validation of a T7 based linear amplification for genomic DNA
Abstract
Background: Genomic maps of transcription factor binding sites and histone modification patterns provide unique insight into the nature of gene regulatory networks and chromatin structure. These systematic studies use microarrays to analyze the composition of DNA isolated by chromatin immunoprecipitation. To obtain quantities sufficient for microarray analysis, the isolated DNA must be amplified. Current protocols use PCR-based approaches to amplify in exponential fashion. However, exponential amplification protocols are highly susceptible to bias. Linear amplification strategies minimize amplification bias and have had a profound impact on mRNA expression analysis. These protocols have yet to be applied to the analysis of genomic DNA due to the lack of a suitable tag such as the polyA tail.
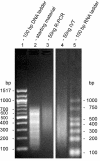
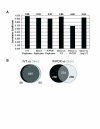
Results: We have developed a novel linear amplification protocol for genomic DNA. Terminal transferase is used to add polyT tails to the ends of DNA fragments. Tail length uniformity is ensured by including a limiting concentration of the terminating nucleotide ddCTP. Second strand synthesis using a T7-polyA primer adapter yields double stranded templates suitable for in vitro transcription (IVT). Using this approach, we are able to amplify as little as 2.5 ng of genomic DNA, while retaining the size distribution of the starting material. In contrast, we find that PCR amplification is biased towards species of greater size. Furthermore, extensive microarray-based analyses reveal that our linear amplification protocol preserves dynamic range and species representation more effectively than a commonly used PCR-based approach.
Conclusion: We present a T7-based linear amplification protocol for genomic DNA. Validation studies and comparisons with existing methods suggest that incorporation of this protocol will reduce amplification bias in genome mapping experiments.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

