Genome-wide analysis of nuclear mRNA export pathways in Drosophila
- PMID: 12743041
- PMCID: PMC155991
- DOI: 10.1093/emboj/cdg233
Genome-wide analysis of nuclear mRNA export pathways in Drosophila
Abstract
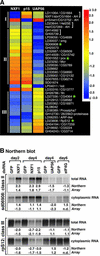
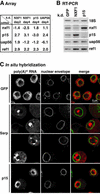
NXF1, p15 and UAP56 are essential nuclear mRNA export factors. The fraction of mRNAs exported by these proteins or via alternative pathways is unknown. We have analyzed the relative abundance of nearly half of the Drosophila transcriptome in the cytoplasm of cells treated with the CRM1 inhibitor leptomycin-B (LMB) or depleted of export factors by RNA interference. While the vast majority of mRNAs were unaffected by LMB, the levels of most mRNAs were significantly reduced in cells depleted of NXF1, p15 or UAP56. The striking similarities of the mRNA expression profiles in NXF1, p15 and UAP56 knockdowns show that these proteins act in the same pathway. The broad effect on mRNA levels observed in these cells indicates that the functioning of this pathway is required for export of most mRNAs. Nonetheless, a set of mRNAs whose export was unaffected by the depletions and some requiring NXF1:p15 but not UAP56 were identified. In addition, our analysis revealed a feedback loop by which a block to mRNA export triggers the upregulation of genes involved in this process.
Figures




References
-
- Alén C., Kent,N.A., Jones,H.S., O’Sullivan,J., Aranda,A. and Proudfoot,N.J. (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell, 10, 1441–1452. - PubMed
-
- Andrulis E.D., Werner,J., Nazarian,A., Erdjument-Bromage,H., Tempst,P. and Lis,J.T. (2002) The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature, 420, 837–841. - PubMed
-
- Gallouzi I.E. and Steitz,J.A. (2001) Delineation of mRNA export pathways by the use of cell-permeable peptides. Science, 294, 1895–1901. - PubMed
-
- Gatfield D., Le Hir,H., Schmitt,C., Braun,I.C., Kocher,T., Wilm,M. and Izaurralde,E. (2001) The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol., 11, 1716–1721. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

