Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains
- PMID: 12743044
- PMCID: PMC155993
- DOI: 10.1093/emboj/cdg235
Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains
Abstract
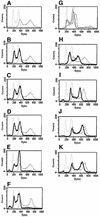
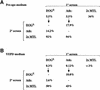
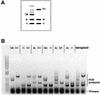
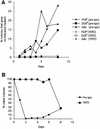
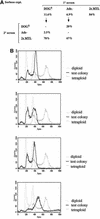
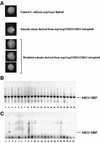
The human pathogenic fungus Candida albicans has traditionally been classified as a diploid, asexual organism. However, mating-competent forms of the organism were recently described that produced tetraploid mating products. In principle, the C.albicans life cycle could be completed via a sexual process, via a parasexual mechanism, or by both mechanisms. Here we describe conditions in which growth of a tetraploid strain of C.albicans on Saccharomyces cerevisiae 'pre-sporulation' medium induced efficient, random chromosome loss in the tetraploid. The products of chromosome loss were often strains that were diploid, or very close to diploid, in DNA content. If they inherited the appropriate MTL (mating-type like) loci, these diploid products were themselves mating competent. Thus, an efficient parasexual cycle can be performed in C.albicans, one that leads to the reassortment of genetic material in this organism. We show that this parasexual cycle-consisting of mating followed by chromosome loss-can be used in the laboratory for simple genetic manipulations in C.albicans.
Figures


References
-
- Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. - PubMed
-
- Edmond M.B., Wallace,S.E., McClish,D.K., Pfaller,M.A., Jones,R.N. and Wenzel,R.P. (1999) Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis., 29, 239–244. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

