Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication
- PMID: 12743046
- PMCID: PMC155996
- DOI: 10.1093/emboj/cdg238
Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication
Abstract
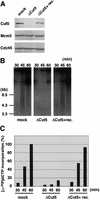
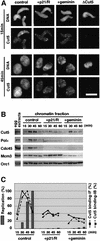
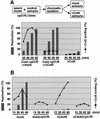
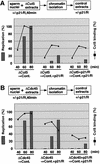
Fission yeast Cut5/Rad4 and its budding yeast homolog Dpb11 are required for both DNA replication and the S-phase checkpoint. Here, we have investigated the role of the Xenopus homolog of Cut5 in the initiation of DNA replication using Xenopus egg extracts. Xenopus Cut5, which shows sequence similarity to DmMus101 and HsTopBP1, is essential for DNA replication in the egg extracts. It is required for the chromatin binding of Cdc45 and DNA polymerases, but not for the formation of pre-replicative complexes or the elongation stage of DNA replication. The chromatin binding of Cut5 consists of two distinct modes. S-phase cyclin-dependent kinase (S-CDK)-independent binding is sufficient for DNA replication while S-CDK-dependent binding is dispensable. Further, S-CDK acts after the chromatin binding of Cut5 and before the binding of Cdc45. These results demonstrate that the chromatin binding of Cut5 is required for the action of S-CDK, which in turn triggers the formation of pre-initiation complexes of DNA replication.
Figures




Similar articles
-
A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication.Genes Dev. 2003 May 1;17(9):1141-52. doi: 10.1101/gad.1070003. Genes Dev. 2003. PMID: 12730133 Free PMC article.
-
Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk.EMBO J. 1998 Oct 1;17(19):5699-707. doi: 10.1093/emboj/17.19.5699. EMBO J. 1998. PMID: 9755170 Free PMC article.
-
Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading.Genes Dev. 2000 Jun 15;14(12):1528-40. Genes Dev. 2000. PMID: 10859170 Free PMC article.
-
Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication.Curr Opin Cell Biol. 2010 Dec;22(6):766-71. doi: 10.1016/j.ceb.2010.07.015. Epub 2010 Aug 20. Curr Opin Cell Biol. 2010. PMID: 20728327 Review.
-
The DNA replication licensing system.Cancer Surv. 1997;29:75-90. Cancer Surv. 1997. PMID: 9338097 Review.
Cited by
-
Resection of DNA double-strand breaks activates Mre11-Rad50-Nbs1- and Rad9-Hus1-Rad1-dependent mechanisms that redundantly promote ATR checkpoint activation and end processing in Xenopus egg extracts.Nucleic Acids Res. 2024 Apr 12;52(6):3146-3163. doi: 10.1093/nar/gkae082. Nucleic Acids Res. 2024. PMID: 38349040 Free PMC article.
-
ATRIP from TopBP1 to ATR--in vitro activation of a DNA damage checkpoint.Proc Natl Acad Sci U S A. 2010 Aug 3;107(31):13561-2. doi: 10.1073/pnas.1008909107. Epub 2010 Jul 26. Proc Natl Acad Sci U S A. 2010. PMID: 20660767 Free PMC article. No abstract available.
-
Eukaryotic DNA replication origins: many choices for appropriate answers.Nat Rev Mol Cell Biol. 2010 Oct;11(10):728-38. doi: 10.1038/nrm2976. Nat Rev Mol Cell Biol. 2010. PMID: 20861881 Review.
-
TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival.Genes Dev. 2004 Mar 15;18(6):673-86. doi: 10.1101/gad.1180204. Genes Dev. 2004. PMID: 15075294 Free PMC article.
-
The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation.Mol Cell Biol. 2010 Mar;30(6):1495-507. doi: 10.1128/MCB.00710-09. Epub 2010 Jan 11. Mol Cell Biol. 2010. PMID: 20065034 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

