Specificity of short interfering RNA determined through gene expression signatures
- PMID: 12746500
- PMCID: PMC164449
- DOI: 10.1073/pnas.1131959100
Specificity of short interfering RNA determined through gene expression signatures
Abstract
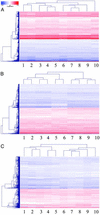
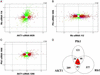
Short interfering RNA (siRNA) is widely used for studying gene function and holds great promise as a tool for validating drug targets and treating disease. A critical assumption in these applications is that the effect of siRNA on cells is specific, i.e., limited to the specific knockdown of the target gene. In this article, we characterize the specificity of siRNA by applying gene expression profiling. Several siRNAs were designed against different regions of the same target gene for three different targets. Their effects on cells were compared by using DNA microarrays to generate gene expression signatures. When the siRNA design and transfection conditions were optimized, the signatures for different siRNAs against the same target were shown to correlate very closely, whereas the signatures for different genes revealed no correlation. These results indicate that siRNA is a highly specific tool for targeted gene knockdown, establishing siRNA-mediated gene silencing as a reliable approach for large-scale screening of gene function and drug target validation.
Figures



Comment in
-
The specifics of small interfering RNA specificity.Proc Natl Acad Sci U S A. 2003 May 27;100(11):6289-91. doi: 10.1073/pnas.1232238100. Epub 2003 May 16. Proc Natl Acad Sci U S A. 2003. PMID: 12754379 Free PMC article. No abstract available.
References
-
- McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 10, 737–747. - PubMed
-
- McManus, M., Haines, B., Dillon, C. P., Whitehurst, C. E., van Parijs, L., Chen, J. & Sharp, P. A. (2002) J. Immunol. 169, 5754–5760. - PubMed
-
- Prasanth, S. G., Prasanth, K. V. & Stillman, B. (2002) Science 297, 1026–1031. - PubMed
-
- DiTullio, R. A., Mochan, T. A., Venere, M., Bartkova, J., Sehested, M., Bartek, J. & Halazonetis, T. D. (2002) Nat. Cell Biol. 4, 998–1002. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

