Microarray analysis: a novel research tool for cardiovascular scientists and physicians
- PMID: 12748210
- PMCID: PMC1767682
- DOI: 10.1136/heart.89.6.597
Microarray analysis: a novel research tool for cardiovascular scientists and physicians
Abstract
The massive increase in information on the human DNA sequence and the development of new technologies will have a profound impact on the diagnosis and treatment of cardiovascular diseases. The microarray is a micro-hybridisation based assay. The filter, called microchip or chip, is a special kind of membrane in which are spotted several thousands of oligonucleotides of cDNA fragments coding for known genes or expressed sequence tags. The resulting hybridisation signal on the chip is analysed by a fluorescent scanner and processed with a software package utilising the information on the oligonucleotide or cDNA map of the chip to generate a list of relative gene expression. Microarray technology can be used for many different purposes, most prominently to measure differential gene expression, variations in gene sequence (by analysing the genome of mutant phenotypes), or more recently, the entire binding site for transcription factors. Measurements of gene expression have the advantage of providing all available sequence information for any given experimental design and data interpretation in pursuit of biological understanding. This research tool will contribute to radically changing our understanding of cardiovascular diseases.
Figures
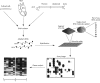
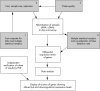
References
-
- Chese M, Yang R, Hubbell E, et al. Accessing genetic information with high-density DNA array. Science 1996;224:610–14. - PubMed
-
- Schena M, Shalson D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270:467–70. - PubMed
-
- Gray DE, Case-Green SC, Fell TS, et al. Ellipsometric and interferometric characterization of DNA probe immobilized on a combinatorial array. Langmuir 1997;13:2833–42.
-
- Barlund M, Monni O, Kononen J, et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 2000;60:5340–4. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
