Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor
- PMID: 12748276
- PMCID: PMC155231
- DOI: 10.1128/MCB.23.11.3707-3720.2003
Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor
Abstract
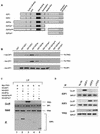
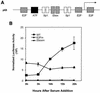
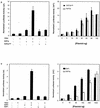
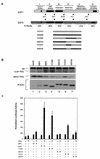
Various studies have demonstrated a role for E2F proteins in the control of transcription of genes involved in DNA replication, cell cycle progression, and cell fate determination. Although it is clear that the functions of the E2F proteins overlap, there is also evidence for specific roles for individual E2F proteins in the control of apoptosis and cell proliferation. Investigating protein interactions that might provide a mechanistic basis for the specificity of E2F function, we identified the E-box binding factor TFE3 as an E2F3-specific partner. We also show that this interaction is dependent on the marked box domain of E2F3. We provide evidence for a role for TFE3 in the synergistic activation of the p68 subunit gene of DNA polymerase alpha together with E2F3, again dependent on the E2F3 marked box domain. Chromatin immunoprecipitation assays showed that TFE3 and E2F3 were bound to the p68 promoter in vivo and that the interaction of either E2F3 or TFE3 with the promoter was facilitated by the presence of both proteins. In contrast, neither E2F1 nor E2F2 interacted with the p68 promoter under these conditions. We propose that the physical interaction of TFE3 and E2F3 facilitates transcriptional activation of the p68 gene and provides strong evidence for the specificity of E2F function.
Figures




Similar articles
-
Combinatorial gene control involving E2F and E Box family members.EMBO J. 2004 Mar 24;23(6):1336-47. doi: 10.1038/sj.emboj.7600134. Epub 2004 Mar 4. EMBO J. 2004. PMID: 15014447 Free PMC article.
-
Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function.EMBO J. 2002 Nov 1;21(21):5775-86. doi: 10.1093/emboj/cdf577. EMBO J. 2002. PMID: 12411495 Free PMC article.
-
Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F.Mol Cell Biol. 1996 Apr;16(4):1659-67. doi: 10.1128/MCB.16.4.1659. Mol Cell Biol. 1996. PMID: 8657141 Free PMC article.
-
Divergent siblings: E2F2 and E2F4 but not E2F1 and E2F3 induce DNA synthesis in cardiomyocytes without activation of apoptosis.Circ Res. 2005 Mar 18;96(5):509-17. doi: 10.1161/01.RES.0000159705.17322.57. Epub 2005 Feb 17. Circ Res. 2005. PMID: 15718499
-
The basic helix-loop-helix-zipper domain of TFE3 mediates enhancer-promoter interaction.Mol Cell Biol. 1994 Dec;14(12):7704-16. doi: 10.1128/mcb.14.12.7704-7716.1994. Mol Cell Biol. 1994. PMID: 7969114 Free PMC article.
Cited by
-
Distinctions in the specificity of E2F function revealed by gene expression signatures.Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):15948-53. doi: 10.1073/pnas.0504300102. Epub 2005 Oct 25. Proc Natl Acad Sci U S A. 2005. PMID: 16249342 Free PMC article.
-
A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription.Genes Dev. 2004 Dec 1;18(23):2941-51. doi: 10.1101/gad.1239304. Genes Dev. 2004. PMID: 15574595 Free PMC article.
-
The E2F family: specific functions and overlapping interests.EMBO J. 2004 Dec 8;23(24):4709-16. doi: 10.1038/sj.emboj.7600481. Epub 2004 Nov 11. EMBO J. 2004. PMID: 15538380 Free PMC article. Review.
-
E2F1-Mediated Induction of NFYB Attenuates Apoptosis via Joint Regulation of a Pro-Survival Transcriptional Program.PLoS One. 2015 Jun 3;10(6):e0127951. doi: 10.1371/journal.pone.0127951. eCollection 2015. PLoS One. 2015. PMID: 26039627 Free PMC article.
-
E2Fs link the control of G1/S and G2/M transcription.EMBO J. 2004 Nov 24;23(23):4615-26. doi: 10.1038/sj.emboj.7600459. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510213 Free PMC article.
References
-
- Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. - PubMed
-
- Brodin, G., A. Hgren, C.-H. Dijke, and R. Heuchel. 2000. Efficient TGF-β induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J. Biol. Chem. 275:29023-29030. - PubMed
-
- Cartwright, P., H. Muller, C. Wagener, K. Holm, and K. Helin. 1998. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17:611-623. - PubMed
-
- Dahl, R., B. Wani, and M. J. Hayman. 1998. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 26:1579-1586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
