Spatial and temporal cellular responses to single-strand breaks in human cells
- PMID: 12748298
- PMCID: PMC155230
- DOI: 10.1128/MCB.23.11.3974-3981.2003
Spatial and temporal cellular responses to single-strand breaks in human cells
Erratum in
- Mol Cell Biol. 2003 Aug;23(15):5472
Abstract
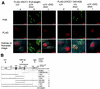
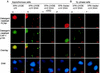
DNA single-strand breaks (SSB) are one of the most frequent DNA lesions produced by reactive oxygen species and during DNA metabolism, but the analysis of cellular responses to SSB remains difficult due to the lack of an experimental method to produce SSB alone in cells. By using human cells expressing a foreign UV damage endonuclease (UVDE) and irradiating the cells with UV through tiny pores in membrane filters, we created SSB in restricted areas in the nucleus by the immediate action of UVDE on UV-induced DNA lesions. Cellular responses to the SSB were characterized by using antibodies and fluorescence microscopy. Upon UV irradiation, poly(ADP-ribose) synthesis occurred immediately in the irradiated area. Simultaneously, but dependent on poly(ADP-ribosyl)ation, XRCC1 was translocated from throughout the nucleus, including nucleoli, to the SSB. The BRCT1 domain of XRCC1 protein was indispensable for its poly(ADP-ribose)-dependent recruitment to the SSB. Proliferating cell nuclear antigen and the p150 subunit of chromatin assembly factor 1 also accumulated at the SSB in a detergent-resistant form, which was significantly reduced by inhibition of poly(ADP-ribose) synthesis. Our results show the importance of poly(ADP-ribosyl)ation in sequential cellular responses to SSB.
Figures


Similar articles
-
The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function.Nucleic Acids Res. 2015 Aug 18;43(14):6934-44. doi: 10.1093/nar/gkv623. Epub 2015 Jun 29. Nucleic Acids Res. 2015. PMID: 26130715 Free PMC article.
-
Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions.Biochem J. 1999 Sep 1;342 ( Pt 2)(Pt 2):249-68. Biochem J. 1999. PMID: 10455009 Free PMC article. Review.
-
Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells.Nucleic Acids Res. 2007;35(22):7665-75. doi: 10.1093/nar/gkm933. Epub 2007 Nov 3. Nucleic Acids Res. 2007. PMID: 17982172 Free PMC article.
-
Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses.DNA Repair (Amst). 2013 Sep;12(9):774-85. doi: 10.1016/j.dnarep.2013.06.004. Epub 2013 Jul 18. DNA Repair (Amst). 2013. PMID: 23871146 Free PMC article.
-
Poly(ADP-ribosyl)ation, DNA strand breaks, chromatin and cancer.Toxicol Lett. 1993 Apr;67(1-3):129-50. doi: 10.1016/0378-4274(93)90051-x. Toxicol Lett. 1993. PMID: 8451755 Review.
Cited by
-
PARP Inhibitors as Monotherapy in Daily Practice for Advanced Prostate Cancers.J Clin Med. 2022 Mar 21;11(6):1734. doi: 10.3390/jcm11061734. J Clin Med. 2022. PMID: 35330059 Free PMC article. Review.
-
PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways.Nucleic Acids Res. 2006;34(21):6170-82. doi: 10.1093/nar/gkl840. Epub 2006 Nov 6. Nucleic Acids Res. 2006. PMID: 17088286 Free PMC article.
-
XRCC1 is required for DNA single-strand break repair in human cells.Nucleic Acids Res. 2005 May 2;33(8):2512-20. doi: 10.1093/nar/gki543. Print 2005. Nucleic Acids Res. 2005. PMID: 15867196 Free PMC article.
-
Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo.EMBO J. 2003 Oct 1;22(19):5163-74. doi: 10.1093/emboj/cdg478. EMBO J. 2003. PMID: 14517254 Free PMC article.
-
Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells.Nucleic Acids Res. 2009 May;37(9):e68. doi: 10.1093/nar/gkp221. Epub 2009 Apr 7. Nucleic Acids Res. 2009. PMID: 19357094 Free PMC article.
References
-
- Althaus, F. R. 1992. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J. Cell Sci. 102:663-670. - PubMed
-
- Ame, J. C., V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia, and G. de Murcia. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860-17868. - PubMed
-
- Burkle, A. 2001. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays 23:795-806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
