In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum
- PMID: 12748301
- PMCID: PMC155205
- DOI: 10.1128/MCB.23.11.4000-4012.2003
In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum
Abstract
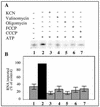
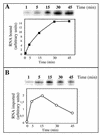
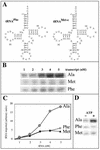
Some of the mitochondrial tRNAs of higher plants are nuclearly encoded and imported into mitochondria. The import of tRNAs encoded in the nucleus has been shown to be essential for proper protein translation within mitochondria of a variety of organisms. Here, we report the development of an in vitro assay for import of nuclearly encoded tRNAs into plant mitochondria. This in vitro system utilizes isolated mitochondria from Solanum tuberosum and synthetic tRNAs transcribed from cloned nuclear tRNA genes. Although incubation of radioactively labeled in vitro-transcribed tRNA(Ala), tRNA(Phe), and tRNA(Met-e) with isolated potato mitochondria resulted in importation, as measured by nuclease protection, the amount of tRNA transcripts protected at saturation was at least five times higher for tRNA(Ala) than for the two other tRNAs. This difference in in vitro saturation levels of import is consistent with the in vivo localization of these tRNAs, since cytosolic tRNA(Ala) is naturally imported into potato mitochondria whereas tRNA(Phe) and tRNA(Met-e) are not. Characterization of in vitro tRNA import requirements indicates that mitochondrial tRNA import proceeds in the absence of any added cytosolic protein fraction, involves at least one protein component on the surface of mitochondria, and requires ATP-dependent step(s) and a membrane potential.
Figures






Similar articles
-
Transfer RNA travels from the cytoplasm to organelles.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):802-17. doi: 10.1002/wrna.93. Epub 2011 Jul 11. Wiley Interdiscip Rev RNA. 2011. PMID: 21976284 Free PMC article. Review.
-
In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei.Mol Cell Biol. 1999 Sep;19(9):6253-9. doi: 10.1128/MCB.19.9.6253. Mol Cell Biol. 1999. PMID: 10454571 Free PMC article.
-
Differential import of nuclear-encoded tRNAGly isoacceptors into solanum Tuberosum mitochondria.Nucleic Acids Res. 1999 May 1;27(9):2037-42. doi: 10.1093/nar/27.9.2037. Nucleic Acids Res. 1999. PMID: 10198438 Free PMC article.
-
Striking differences in mitochondrial tRNA import between different plant species.Mol Gen Genet. 1996 Sep 25;252(4):404-11. doi: 10.1007/BF02173005. Mol Gen Genet. 1996. PMID: 8879241
-
Mechanisms of tRNA import into yeast mitochondria: an overview.Biochimie. 1996;78(6):502-10. doi: 10.1016/0300-9084(96)84756-0. Biochimie. 1996. PMID: 8915539 Review.
Cited by
-
The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18362-7. doi: 10.1073/pnas.0606449103. Epub 2006 Nov 14. Proc Natl Acad Sci U S A. 2006. PMID: 17105808 Free PMC article.
-
Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion.Genes Dev. 2005 Mar 1;19(5):583-92. doi: 10.1101/gad.1269305. Epub 2005 Feb 10. Genes Dev. 2005. PMID: 15706032 Free PMC article.
-
Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria.Curr Genet. 2009 Feb;55(1):1-18. doi: 10.1007/s00294-008-0223-9. Epub 2008 Dec 16. Curr Genet. 2009. PMID: 19083240 Review.
-
Transfer RNA travels from the cytoplasm to organelles.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):802-17. doi: 10.1002/wrna.93. Epub 2011 Jul 11. Wiley Interdiscip Rev RNA. 2011. PMID: 21976284 Free PMC article. Review.
-
Non-coding RNA Regulated Cross-Talk Between Mitochondria and Other Cellular Compartments.Front Cell Dev Biol. 2021 Aug 3;9:688523. doi: 10.3389/fcell.2021.688523. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34414182 Free PMC article. Review.
References
-
- Adhya, S., T. Ghosh, A. Das, S. K. Bera, and S. Mahapatra. 1997. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem. 272:21396-21402. - PubMed
-
- Akama, K., and M. Kashihara. 1996. Plant nuclear tRNAMet genes are ubiquitously interrupted by introns. Plant Mol. Biol. 32:427-434. - PubMed
-
- Aphasizhev, R., U. Karmarkar, and L. Simpson. 1998. Are tRNAs imported into the mitochondria of kinetoplastid protozoa as 5′-extended precursors? Mol. Biochem. Parasitol. 93:73-80. - PubMed
-
- Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25-31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
