Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alphavbeta3
- PMID: 12748384
- PMCID: PMC165870
- DOI: 10.1073/pnas.1131117100
Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alphavbeta3
Abstract
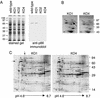
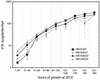
Borrelia burgdorferi, the agent of Lyme disease, expresses several adhesion molecules that are probably required for initial establishment of infection in mammalian hosts, and for colonization of various tissues within the host. The B. burgdorferi outer membrane protein P66 was previously identified as a ligand for beta3-chain integrins by using a variety of biochemical approaches. Although the earlier data suggested that P66 is an adhesin that mediates B. burgdorferi attachment to beta3-chain integrins, lack of genetic systems in B. burgdorferi precluded definitive demonstration of a role for P66 in beta3 integrin attachment by intact borreliae. Recent advances in the genetic manipulation of B. burgdorferi have now made possible the targeted disruption of the p66 gene. Mutants in p66 show dramatically reduced attachment to integrin alphavbeta3. This is, to our knowledge, the first description of the targeted disruption of a candidate B. burgdorferi virulence factor with a known biochemical function that can be quantified, and demonstrates the importance of B. burgdorferi P66 in the attachment of this pathogenic spirochete to a human cell-surface receptor.
Figures

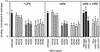

Similar articles
-
Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete.Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7307-12. doi: 10.1073/pnas.1231043100. Epub 2003 May 28. Proc Natl Acad Sci U S A. 2003. PMID: 12773620 Free PMC article.
-
Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library.Mol Microbiol. 1999 Dec;34(5):926-40. doi: 10.1046/j.1365-2958.1999.01654.x. Mol Microbiol. 1999. PMID: 10594819
-
Integrin binding by Borrelia burgdorferi P66 facilitates dissemination but is not required for infectivity.Cell Microbiol. 2015 Jul;17(7):1021-36. doi: 10.1111/cmi.12418. Epub 2015 Feb 16. Cell Microbiol. 2015. PMID: 25604835 Free PMC article.
-
Adhesion mechanisms of Borrelia burgdorferi.Adv Exp Med Biol. 2011;715:35-49. doi: 10.1007/978-94-007-0940-9_3. Adv Exp Med Biol. 2011. PMID: 21557056 Free PMC article. Review.
-
Borrelia burgdorferi and its tropisms for adhesion molecules in the joint.Curr Opin Rheumatol. 2002 Jul;14(4):394-8. doi: 10.1097/00002281-200207000-00010. Curr Opin Rheumatol. 2002. PMID: 12118173 Review.
Cited by
-
Development and validation of systems for genetic manipulation of the Old World tick-borne relapsing fever spirochete, Borrelia duttonii.PLoS Negl Trop Dis. 2024 Jul 22;18(7):e0012348. doi: 10.1371/journal.pntd.0012348. eCollection 2024 Jul. PLoS Negl Trop Dis. 2024. PMID: 39038047 Free PMC article.
-
Leptospiral outer membrane protein microarray, a novel approach to identification of host ligand-binding proteins.J Bacteriol. 2012 Nov;194(22):6074-87. doi: 10.1128/JB.01119-12. Epub 2012 Sep 7. J Bacteriol. 2012. PMID: 22961849 Free PMC article.
-
Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection.Infect Immun. 2008 Dec;76(12):5694-705. doi: 10.1128/IAI.00690-08. Epub 2008 Sep 22. Infect Immun. 2008. PMID: 18809667 Free PMC article.
-
Identification of amino acid domains of Borrelia burgdorferi P66 that are surface exposed and important for localization, oligomerization, and porin function of the protein.Front Cell Infect Microbiol. 2022 Sep 23;12:991689. doi: 10.3389/fcimb.2022.991689. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36211976 Free PMC article.
-
The early dissemination defect attributed to disruption of decorin-binding proteins is abolished in chronic murine Lyme borreliosis.Infect Immun. 2013 May;81(5):1663-73. doi: 10.1128/IAI.01359-12. Epub 2013 Mar 4. Infect Immun. 2013. PMID: 23460518 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

