Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin
- PMID: 12750471
- PMCID: PMC164452
- DOI: 10.1073/pnas.1131966100
Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin
Erratum in
- Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15273
Abstract
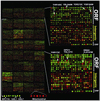
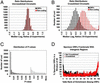
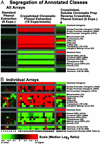
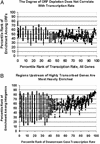
Epigenetic modifications of chromatin serve an important role in regulating the expression and accessibility of genomic DNA. We report here a genomewide approach for fractionating yeast chromatin into two functionally distinct parts, one containing RNA polymerase II transcribed sequences, and the other comprising noncoding sequences and genes transcribed by RNA polymerases I and III. Noncoding regions could be further fractionated into promoters and segments lacking promoters. The observed separations were apparently based on differential crosslinking efficiency of chromatin in different genomic regions. The results reveal a genomewide molecular mechanism for marking promoters and genomic regions that have a license to be transcribed by RNA polymerase II, a previously unrecognized level of genomic complexity that may exist in all eukaryotes. Our approach has broad potential use as a tool for genome annotation and for the characterization of global changes in chromatin structure that accompany different genetic, environmental, and disease states.
Figures
References
-
- Richards, E. J. & Elgin, S. C. (2002) Cell 108, 489–500. - PubMed
-
- Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R. E. & Kouzarides, T. (2001) Nature 412, 561–565. - PubMed
-
- van Leeuwen, F. & Gottschling, D. E. (2002) Curr. Opin. Cell Biol. 14, 756–762. - PubMed
-
- Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

