Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide
- PMID: 12754224
- PMCID: PMC155371
- DOI: 10.1128/JB.185.11.3270-3277.2003
Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide
Abstract
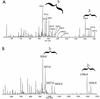
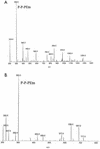
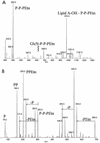
A gene, NMB1638, with homology to the recently characterized gene encoding a phosphoethanolamine transferase, lpt-3, has been identified from the Neisseria meningitidis genome sequence and was found to be present in all meningococcal strains examined. Homology comparison with other database sequences would suggest that NMB1638 and lpt-3 represent genes coding for members of a family of proteins of related function identified in a wide range of gram-negative species of bacteria. When grown and isolated under appropriate conditions, N. meningitidis elaborated lipopolysaccharide (LPS) containing a lipid A that was characteristically phosphorylated with multiple phosphate and phosphoethanolamine residues. In all meningococcal strains examined, each lipid A species contained the basal diphosphorylated species, wherein a phosphate group is attached to each glucosamine residue. Also elaborated within the population of LPS molecules are a variety of "phosphoforms" that contain either an additional phosphate residue, an additional phosphoethanolamine residue, additional phosphate and phosphoethanolamine residues, or an additional phosphate and two phosphoethanolamine residues in the lipid A. Mass spectroscopic analyses of LPS from three strains in which NMB1638 had been inactivated by a specific mutation indicated that there were no phosphoethanolamine residues included in the lipid A region of the LPS and that there was no further phosphorylation of lipid A beyond one additional phosphate species. We propose that NMB1638 encodes a phosphoethanolamine transferase specific for lipid A and propose naming the gene "lptA," for "LPS phosphoethenolamine transferase for lipid A."
Figures
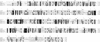






Similar articles
-
The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin.J Mol Biol. 2013 Sep 23;425(18):3389-402. doi: 10.1016/j.jmb.2013.06.029. Epub 2013 Jun 28. J Mol Biol. 2013. PMID: 23810904
-
Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding.Proc Natl Acad Sci U S A. 2017 Feb 28;114(9):2218-2223. doi: 10.1073/pnas.1612927114. Epub 2017 Feb 13. Proc Natl Acad Sci U S A. 2017. PMID: 28193899 Free PMC article.
-
Characterization of Two Novel Lipopolysaccharide Phosphoethanolamine Transferases in Pasteurella multocida and Their Role in Resistance to Cathelicidin-2.Infect Immun. 2017 Oct 18;85(11):e00557-17. doi: 10.1128/IAI.00557-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28874446 Free PMC article.
-
Lipid A Phosphoethanolamine Transferase: Regulation, Structure and Immune Response.J Mol Biol. 2020 Aug 21;432(18):5184-5196. doi: 10.1016/j.jmb.2020.04.022. Epub 2020 Apr 27. J Mol Biol. 2020. PMID: 32353363 Review.
-
[Analyses of the pathogenesis in Neisseria meningitidis].Nihon Saikingaku Zasshi. 2009 Dec;64(2-4):291-301. doi: 10.3412/jsb.64.291. Nihon Saikingaku Zasshi. 2009. PMID: 19628927 Review. Japanese. No abstract available.
Cited by
-
Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine.J Biol Chem. 2012 Aug 24;287(35):29384-96. doi: 10.1074/jbc.M112.380212. Epub 2012 Jul 2. J Biol Chem. 2012. PMID: 22761430 Free PMC article.
-
Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core.J Bacteriol. 2005 May;187(10):3391-9. doi: 10.1128/JB.187.10.3391-3399.2005. J Bacteriol. 2005. PMID: 15866924 Free PMC article.
-
Colistin Sensitivity and Factor H-Binding Protein Expression among Commensal Neisseria Species.mSphere. 2021 Jun 30;6(3):101128msphere0017521. doi: 10.1128/mSphere.00175-21. Epub 2021 Jun 16. mSphere. 2021. PMID: 34133203 Free PMC article.
-
Spread of MCR-3 Colistin Resistance in China: An Epidemiological, Genomic and Mechanistic Study.EBioMedicine. 2018 Aug;34:139-157. doi: 10.1016/j.ebiom.2018.07.027. Epub 2018 Jul 27. EBioMedicine. 2018. PMID: 30061009 Free PMC article.
-
The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica.J Bacteriol. 2004 Jul;186(13):4124-33. doi: 10.1128/JB.186.13.4124-4133.2004. J Bacteriol. 2004. PMID: 15205413 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. J. Wiley and Sons, New York, N.Y.
-
- Cox, A. D., and S. G. Wilkinson. 1991. Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol. Microbiol. 5:641-646. - PubMed
-
- Cox, A. D., J. Li, J.-R. Brisson, E. R. Moxon, and J. C. Richards. 2002. Structural analysis of the lipopolysaccharide from Neisseria meningitidis strain BZ157 galE: localisation of two phosphoethanolamine residues in the inner core oligosaccharide. Carbohydr. Res. 337:1435-1444. - PubMed
-
- Di Fabio, J. L., F. Michon, J. Brisson, and H. J. Jennings. 1990. Structure of L1 and L6 core oligosaccharide epitopes of Neisseria meningitidis. Can. J. Chem. 68:1029-1034.
-
- Gamian, A., M. Beurret, F. Michon, J. R. Brisson, and H. J. Jennings. 1992. Structure of the L2 lipopolysaccharide core oligosaccharides of Neisseria meningitidis. J. Biol. Chem. 267:922-925. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

