How clonal is Staphylococcus aureus?
- PMID: 12754228
- PMCID: PMC155367
- DOI: 10.1128/JB.185.11.3307-3316.2003
How clonal is Staphylococcus aureus?
Abstract
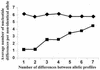
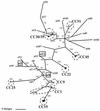
Staphylococcus aureus is an important human pathogen and represents a growing public health burden owing to the emergence and spread of antibiotic-resistant clones, particularly within the hospital environment. Despite this, basic questions about the evolution and population biology of the species, particularly with regard to the extent and impact of homologous recombination, remain unanswered. We address these issues through an analysis of sequence data obtained from the characterization by multilocus sequence typing (MLST) of 334 isolates of S. aureus, recovered from a well-defined population, over a limited time span. We find no significant differences in the distribution of multilocus genotypes between strains isolated from carriers and those from patients with invasive disease; there is, therefore, no evidence from MLST data, which index variation within the stable "core" genome, for the existence of hypervirulent clones of this pathogen. Examination of the sequence changes at MLST loci during clonal diversification shows that point mutations give rise to new alleles at least 15-fold more frequently than does recombination. This contrasts with the naturally transformable species Neisseria meningitidis and Streptococcus pneumoniae, in which alleles change between 5- and 10-fold more frequently by recombination than by mutation. However, phylogenetic analysis suggests that homologous recombination does contribute toward the evolution of this species over the long term. Finally, we note a striking excess of nonsynonymous substitutions in comparisons between isolates belonging to the same clonal complex compared to isolates belonging to different clonal complexes, suggesting that the removal of deleterious mutations by purifying selection may be relatively slow.
Figures


References
-
- Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. Clonal relationships between invasive and carriage Streptococcus pneumoniae, and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis., in press. - PubMed
-
- Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. (Retraction, 295:971, 2002.) - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

