Conserved eukaryotic histone-fold residues substituted into an archaeal histone increase DNA affinity but reduce complex flexibility
- PMID: 12754245
- PMCID: PMC155370
- DOI: 10.1128/JB.185.11.3453-3457.2003
Conserved eukaryotic histone-fold residues substituted into an archaeal histone increase DNA affinity but reduce complex flexibility
Abstract
Although the archaeal and eukaryotic nucleosome core histones evolved from a common ancestor, conserved lysine residues are present at DNA-binding locations in all four eukaryotic histones that are not present in the archaeal histones. Introduction of lysine residues at the corresponding locations into an archaeal histone, HMfB, generated a variant with increased affinity for DNA that formed more compact complexes with DNA. However, these complexes no longer facilitated the circularization of short DNA molecules and had lost the flexibility to wrap DNA alternatively in either a negative or positive supercoil.
Figures

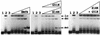
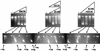
Similar articles
-
Mutational analysis of archaeal histone-DNA interactions.J Mol Biol. 2000 Mar 17;297(1):39-47. doi: 10.1006/jmbi.2000.3546. J Mol Biol. 2000. PMID: 10704305
-
Archaeal histones: structures, stability and DNA binding.Biochem Soc Trans. 2004 Apr;32(Pt 2):227-30. doi: 10.1042/bst0320227. Biochem Soc Trans. 2004. PMID: 15046577 Review.
-
Archaeal histones and the origin of the histone fold.Curr Opin Microbiol. 2006 Oct;9(5):520-5. doi: 10.1016/j.mib.2006.08.003. Epub 2006 Aug 22. Curr Opin Microbiol. 2006. PMID: 16920388
-
Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling.J Biol Chem. 2002 Aug 23;277(34):30879-86. doi: 10.1074/jbc.M203674200. Epub 2002 Jun 10. J Biol Chem. 2002. PMID: 12058041
-
Histones and nucleosomes in Archaea and Eukarya: a comparative analysis.Extremophiles. 1998 Aug;2(3):141-8. doi: 10.1007/s007920050053. Extremophiles. 1998. PMID: 9783158 Review.
Cited by
-
Old cogs, new tricks: the evolution of gene expression in a chromatin context.Nat Rev Genet. 2019 May;20(5):283-297. doi: 10.1038/s41576-019-0105-7. Nat Rev Genet. 2019. PMID: 30886348 Review.
-
Structure and function of archaeal histones.PLoS Genet. 2018 Sep 13;14(9):e1007582. doi: 10.1371/journal.pgen.1007582. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30212449 Free PMC article. Review.
-
Histone variants in archaea and the evolution of combinatorial chromatin complexity.Proc Natl Acad Sci U S A. 2020 Dec 29;117(52):33384-33395. doi: 10.1073/pnas.2007056117. Epub 2020 Dec 7. Proc Natl Acad Sci U S A. 2020. PMID: 33288720 Free PMC article.
-
The hydrophobicity of the H3 histone fold differs from the hydrophobicity of the other three folds.J Mol Evol. 2005 Mar;60(3):354-64. doi: 10.1007/s00239-004-0193-6. J Mol Evol. 2005. PMID: 15871046
References
-
- Bailey, K. A., S. L. Pereira., J. Widom, and J. N. Reeve. 2000. Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution. J. Mol. Biol. 303:25-34. - PubMed
-
- Bailey, K. A., F. Marc, K. Sandman, and J. N. Reeve. 2002. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J. Biol. Chem. 277:9293-9301. - PubMed
-
- Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

