Involvement of multiple signaling pathways in follicular lymphoma transformation: p38-mitogen-activated protein kinase as a target for therapy
- PMID: 12756297
- PMCID: PMC165863
- DOI: 10.1073/pnas.1137463100
Involvement of multiple signaling pathways in follicular lymphoma transformation: p38-mitogen-activated protein kinase as a target for therapy
Abstract
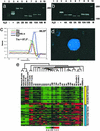
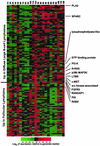
Follicular lymphoma (FL) is the most common form of low-grade non-Hodgkin's lymphoma. Transformation to diffuse large B cell lymphoma (DLBCL) is an important cause of mortality. Using cDNA microarray analysis we identified 113 transformation-associated genes whose expression differed consistently between serial clonally related samples of FL and DLBCL occurring within the same individual. Quantitative RT-PCR validated the microarray results and assigned blinded independent group of 20 FLs, 20 DLBCLs, and five transformed lymphoma-derived cell lines with 100%, 70%, and 100% accuracy, respectively. Notably, growth factor cytokine receptors and p38beta-mitogen-activated protein kinase (MAPK) were differentially expressed in the DLBCLs. Immunohistochemistry of another blinded set of samples demonstrated expression of phosphorylated p38MAPK in 6/6 DLBCLs and 1/5 FLs, but not in benign germinal centers. SB203580 an inhibitor of p38MAPK specifically induced caspase-3-mediated apoptosis in t(14;18)+/p38MAPK+-transformed FL-derived cell lines. Lymphoma growth was also inhibited in SB203580-treated NOD-SCID mice. Our results implicate p38MAPK dysregulation in FL transformation and suggest that molecular targeting of specific elements within this pathway should be explored for transformed FL therapy.
Figures




References
-
- Armitage, J. O. & Weisenburger, D. D. (1998) J. Clin. Oncol. 16, 2780-2795. - PubMed
-
- Horning, S. J. & Rosenberg, S. A. (1984) N. Engl. J. Med. 311, 1471-1475. - PubMed
-
- Rowley, J. D. (1988) J. Clin. Oncol. 6, 919-925. - PubMed
-
- Tsujimoto, Y., Finger, L. R., Yunis, J., Nowell, P. C. & Croce, C. M. (1984) Science 226, 1097-1099. - PubMed
-
- Hockenbery, D., Nunez, G., Milliman, C., Schreiber, R. D. & Korsmeyer, S. J. (1990) Nature 348, 334-336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

