Metabolite-binding RNA domains are present in the genes of eukaryotes
- PMID: 12756322
- PMCID: PMC1370431
- DOI: 10.1261/rna.5090103
Metabolite-binding RNA domains are present in the genes of eukaryotes
Abstract
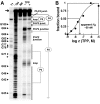
Genetic control by metabolite-binding mRNAs is widespread in prokaryotes. These riboswitches are typically located in noncoding regions of mRNA, where they selectively bind their target compound and subsequently modulate gene expression. We have identified mRNA elements in fungi and in plants that match the consensus sequence and structure of thiamine pyrophosphate-binding domains of prokaryotes. In Arabidopsis, the consensus motif resides in the 3'-UTR of a thiamine biosynthetic gene, and the isolated RNA domain binds the corresponding coenzyme in vitro. These results suggest that metabolite-binding mRNAs are possibly involved in eukaryotic gene regulation and that some riboswitches might be representatives of an ancient form of genetic control.
Figures


References
-
- Begley, T.P., Downs, D.M., Ealick, S.E., McLafferty, F.W., Van Loon, A.P., Tayla, S., Campobasso, N., Chiu, H.J., Kinsland, C., Reddick, J.J., et al. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171: 293–300. - PubMed
-
- Gelfand, M.S., Mironov, A.A., Jomantas, J., Kozlov, Y.I., and Perumov, D.A. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15: 439–442. - PubMed
-
- Hesselberth, J.R., Robertson, M.P., Knudsen, S.M., and Ellington, A.D. 2003. Simultaneous detection of diverse analytes with aptazyme ligase array. Anal. Biochem. 312: 106–112. - PubMed
-
- Jeffares, D.C., Poole, A.M., and Penny, D. 1998. Relics from the RNA world. J. Mol. Evol. 46: 18–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
