The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast
- PMID: 12756324
- PMCID: PMC1370433
- DOI: 10.1261/rna.5100903
The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast
Abstract
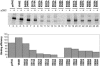
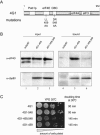
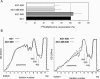
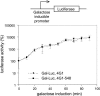
In eukaryotes, the m(7)GpppN cap structure is added to all nascent RNA polymerase II transcripts, and serves important functions at multiple steps of RNA metabolism. The predominantly nuclear cap-binding complex (CBC) binds to the cap during RNA synthesis. The predominantly cytoplasmic eukaryotic initiation factor 4F (eIF4F) is thought to replace CBC after export of mature mRNA to the cytoplasm, and mediates the bulk of cellular translation. Yeast as well as mammalian CBC interacts in vitro with eIF4G, a subunit of eIF4F. In this work, we investigate a potential role of this interaction during translation in yeast. We identify a mutation (DR548/9AA) in Tif4631p, one of two isoforms of yeast eIF4G, that abolishes its binding to CBC. Cells expressing this mutant protein as the sole source of eIF4G grow at wild-type rates, and bulk cellular translation, as assessed by metabolic labeling and polysome profile analysis, is unchanged. Importantly, we find that the DR548/9AA mutation neither diminishes nor delays the translation of newly induced reporter mRNA. Finally, microarray analysis reveals marked transcriptome alterations in CBC subunit deletion strains, whereas eIF4G point mutants have essentially a wild-type transcriptome composition. Collectively, these data suggest that in yeast, the phenotypic consequences of CBC deletions are separable from its interaction with eIF4G, and that the CBC-eIF4G interaction is dispensable for a potential "pioneering round" of translation in yeast.
Figures
References
-
- Colot, H.V., Stutz, F., and Rosbash, M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes & Dev. 10: 1699–1708. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
