Substrate specificity and reaction kinetics of an X-motif ribozyme
- PMID: 12756327
- PMCID: PMC1370436
- DOI: 10.1261/rna.2600503
Substrate specificity and reaction kinetics of an X-motif ribozyme
Abstract
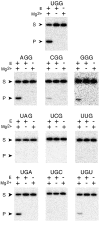
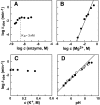
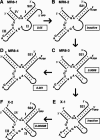
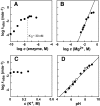
The X-motif is an in vitro-selected ribozyme that catalyzes RNA cleavage by an internal phosphoester transfer reaction. This ribozyme class is distinguished by the fact that it emerged as the dominant clone among at least 12 different classes of ribozymes when in vitro selection was conducted to favor the isolation of high-speed catalysts. We have examined the structural and kinetic properties of the X-motif in order to provide a framework for its application as an RNA-cleaving agent and to explore how this ribozyme catalyzes phosphoester transfer with a predicted rate constant that is similar to those exhibited by the four natural self-cleaving ribozymes. The secondary structure of the X-motif includes four stem elements that form a central unpaired junction. In a bimolecular format, two of these base-paired arms define the substrate specificity of the ribozyme and can be changed to target different RNAs for cleavage. The requirements for nucleotide identity at the cleavage site are GD, where D = G, A, or U and cleavage occurs between the two nucleotides. The ribozyme has an absolute requirement for a divalent cation cofactor and exhibits kinetic behavior that is consistent with the obligate binding of at least two metal ions.
Figures
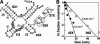
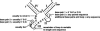
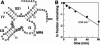
Similar articles
-
Biochemical analysis of hatchet self-cleaving ribozymes.RNA. 2015 Nov;21(11):1845-51. doi: 10.1261/rna.052522.115. Epub 2015 Sep 18. RNA. 2015. PMID: 26385510 Free PMC article.
-
Structural diversity of self-cleaving ribozymes.Proc Natl Acad Sci U S A. 2000 May 23;97(11):5784-9. doi: 10.1073/pnas.97.11.5784. Proc Natl Acad Sci U S A. 2000. PMID: 10823936 Free PMC article.
-
Biochemical analysis of pistol self-cleaving ribozymes.RNA. 2015 Nov;21(11):1852-8. doi: 10.1261/rna.052514.115. Epub 2015 Sep 18. RNA. 2015. PMID: 26385507 Free PMC article.
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
-
Ribozymes: from mechanistic studies to applications in vivo.J Biochem. 1995 Aug;118(2):251-8. doi: 10.1093/oxfordjournals.jbchem.a124899. J Biochem. 1995. PMID: 8543555 Review.
Cited by
-
Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions.RNA. 2015 Mar;21(3):458-69. doi: 10.1261/rna.048033.114. Epub 2015 Jan 20. RNA. 2015. PMID: 25605963 Free PMC article.
-
A common speed limit for RNA-cleaving ribozymes and deoxyribozymes.RNA. 2003 Aug;9(8):949-57. doi: 10.1261/rna.5670703. RNA. 2003. PMID: 12869706 Free PMC article.
-
iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications.Nucleic Acids Res. 2016 Apr 7;44(6):2491-500. doi: 10.1093/nar/gkw083. Epub 2016 Mar 1. Nucleic Acids Res. 2016. PMID: 26932363 Free PMC article.
-
The importance of peripheral sequences in determining the metal selectivity of an in vitro-selected Co(2+) -dependent DNAzyme.Chembiochem. 2012 Feb 13;13(3):381-91. doi: 10.1002/cbic.201100724. Epub 2012 Jan 17. Chembiochem. 2012. PMID: 22250000 Free PMC article.
-
Natural and artificial RNAs occupy the same restricted region of sequence space.RNA. 2010 Feb;16(2):280-9. doi: 10.1261/rna.1923210. Epub 2009 Dec 23. RNA. 2010. PMID: 20032164 Free PMC article.
References
-
- Doherty, E.A. and Doudna, J.A. 2000. Ribozyme structure and mechanisms. Annu. Rev. Biochem. 69: 597–615. - PubMed
-
- Forster, A.C. and Symons, R.H. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49: 211–220. - PubMed
-
- Ishizaka, M., Oshima, Y., and Tani, T. 1995. Isolation of active ribozymes from an RNA pool of random sequences using an anchored substrate RNA. Biochem. Biophys. Res. Commun. 214: 403–409. - PubMed
-
- James, H.A. and Gibson, I. 1998. The therapeutic potential of ribozymes. Blood 91: 371–382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
