A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya
- PMID: 12756329
- PMCID: PMC1370438
- DOI: 10.1261/rna.5230603
A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya
Abstract
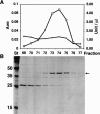
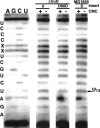
Putative pseudouridine synthase genes are members of a class consisting of four subgroups that possess characteristic amino acid sequence motifs. These genes have been found in all organisms sequenced to date. In Escherichia coli, 10 such genes have been identified, and the 10 synthase gene products have been shown to function in making all of the pseudouridines found in tRNA and ribosomal RNA except for tRNA(Glu) pseudouridine13. In this work, a protein able to make this pseudouridine was purified by standard biochemical procedures. Amino-terminal sequencing of the isolated protein identified the synthase as YgbO. Deletion of the ygbO gene caused the loss of tRNA(Glu) pseudouridine13 and plasmid-borne restoration of the structural gene restored pseudouridine13. Reaction of the overexpressed gene product, renamed TruD, with a tRNA(Glu) transcript made in vitro also yielded only pseudouridine13. A search of the database detected 58 homologs of TruD spanning all three phylogenetic domains, including ancient organisms. Thus, we have identified a new wide-spread class of pseudouridine synthase with no sequence homology to the previously known four subgroups. The only completely conserved sequence motif in all 59 organisms that contained aspartate was GXKD, in motif II. This aspartate was essential for in vitro activity.
Figures


Similar articles
-
Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli.RNA. 2001 Nov;7(11):1603-15. RNA. 2001. PMID: 11720289 Free PMC article.
-
A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli.RNA. 1998 Nov;4(11):1407-17. doi: 10.1017/s1355838298981146. RNA. 1998. PMID: 9814761 Free PMC article.
-
A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA(phe).RNA. 1995 Jun;1(4):437-48. RNA. 1995. PMID: 7493321 Free PMC article.
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
-
[Pseudouridine synthases--enzymes introducing the most abundant modified nucleoside in nucleic acids--pseudouridine].Postepy Biochem. 2001;47(3):232-42. Postepy Biochem. 2001. PMID: 11908092 Review. Polish. No abstract available.
Cited by
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
-
A pseudouridine synthase homologue is critical to cellular differentiation in Toxoplasma gondii.Eukaryot Cell. 2009 Mar;8(3):398-409. doi: 10.1128/EC.00329-08. Epub 2009 Jan 5. Eukaryot Cell. 2009. PMID: 19124578 Free PMC article.
-
Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation.World J Biol Chem. 2014 Nov 26;5(4):398-408. doi: 10.4331/wjbc.v5.i4.398. World J Biol Chem. 2014. PMID: 25426264 Free PMC article. Review.
-
Matching tRNA modifications in humans to their known and predicted enzymes.Nucleic Acids Res. 2019 Mar 18;47(5):2143-2159. doi: 10.1093/nar/gkz011. Nucleic Acids Res. 2019. PMID: 30698754 Free PMC article.
-
The Evolution of Substrate Specificity by tRNA Modification Enzymes.Enzymes. 2017;41:51-88. doi: 10.1016/bs.enz.2017.03.002. Epub 2017 Apr 26. Enzymes. 2017. PMID: 28601226 Free PMC article. Review.
References
-
- Aravind, L. and Koonin, E.V. 1999. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48: 291–301. - PubMed
-
- ———. 2001. THUMP-a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 26: 215–217. - PubMed
-
- Anantharaman, V., Koonin, E.V., and Aravind, L. 2001. TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol. Lett. 19: 215–221. - PubMed
-
- Argaman, L., Hershberg, R., Vogel, J., Bejerano, G., Wagner, E.G., Margalit, H., and Altuvia, S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11: 941–950. - PubMed
-
- Arluison, V., Hountondji, C., Robert, B., and Grosjean, H. 1998. Transfer RNA-pseudouridine synthetase Pus1 of Saccharomyces cerevisiae contains one atom of zinc essential for its native conformation and tRNA recognition. Biochemistry 37: 7268–7276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
