Comparison of bipA alleles within and across Bordetella species
- PMID: 12761081
- PMCID: PMC155730
- DOI: 10.1128/IAI.71.6.3043-3052.2003
Comparison of bipA alleles within and across Bordetella species
Abstract
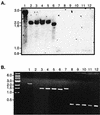
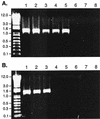
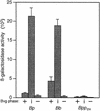
The Bordetella BvgAS signal transduction system controls the expression of at least three phenotypic phases, the Bvg(+) or virulent phase, the Bvg(-) or avirulent phase, and the Bvg(i) or Bvg intermediate phase, which has been hypothesized to be important for transmission. bipA, the first identified Bvg(i)-phase gene, encodes a protein with similarity to the well-characterized bacterial adhesins intimin and invasin. Proteins encoded by the bipA genes present in Bordetella pertussis Tohama I and Bordetella bronchiseptica RB50 differ in the number of 90-amino-acid repeats which they possess and in the sequence of the C-terminal domain. To investigate the possibility that bipA alleles segregate according to host specificity and to gain insight into the role of BipA and the Bvg(i) phase in the Bordetella infectious cycle, we compared bipA alleles across members of the B. bronchiseptica cluster, which includes both human-infective (B. pertussis and B. parapertussis(hu)) and non-human-infective (B. bronchiseptica and B. parapertussis(ov)) strains. bipA genes were present in most, but not all, strains. All bipA genes present in B. bronchiseptica strains were identical to bipA of RB50 (at least with regard to the DNA sequence of the 3' C-terminal-domain-encoding region, the number of 90-amino-acid repeats encoded, and expression patterns). Although all bipA genes present in the other Bordetella strains were identical in the 3' C-terminal-domain-encoding region to bipA of B. pertussis Tohama I, they varied in the number of 90-amino-acid repeats that they encoded and in expression level. Notably, the genes present in B. parapertussis(hu) strains were pseudogenes, and the genes present in B. parapertussis(ov) strains were expressed at significantly reduced levels compared with the levels in B. pertussis and B. bronchiseptica strains. Our results indicate that there is a correlation between specific bipA alleles and specific hosts. They also support the hypothesis that both horizontal gene transfer and fine-tuning of gene expression patterns contribute to the evolution of host adaptation in lineages of the B. bronchiseptica cluster.
Figures




References
-
- Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. - PubMed
-
- Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 19:37-52. - PubMed
-
- Arico, B., R. Gross, J. Smida, and R. Rappuoli. 1987. Evolutionary relationships in the genus Bordetella. Mol. Microbiol. 1:301-308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

