Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals
- PMID: 12761121
- PMCID: PMC155701
- DOI: 10.1128/IAI.71.6.3371-3383.2003
Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals
Abstract
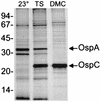
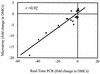
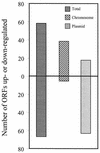
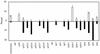
Lyme disease is a tick-borne infection that can lead to chronic, debilitating problems if not recognized or treated appropriately. Borrelia burgdorferi, the causative agent of Lyme disease, is maintained in nature by a complex enzootic cycle involving Ixodes ticks and mammalian hosts. Many previous studies support the notion that B. burgdorferi differentially expresses numerous genes and proteins to help it adapt to growth in the mammalian host. In this regard, several studies have utilized a dialysis membrane chamber (DMC) cultivation system to generate "mammalian host-adapted" spirochetes for the identification of genes selectively expressed during mammalian infection. Here, we have exploited the DMC cultivation system in conjunction with microarray technology to examine the global changes in gene expression that occur in the mammalian host. To identify genes regulated by only mammal-specific signals and not by temperature, borrelial microarrays were hybridized with cDNA generated either from organisms temperature shifted in vitro from 23 degrees C to 37 degrees C or from organisms cultivated by using the DMC model system. Statistical analyses of the combined data sets revealed that 125 genes were expressed at significantly different levels in the mammalian host, with almost equivalent numbers of genes being up- or down-regulated by B. burgdorferi within DMCs compared to those undergoing temperature shift. Interestingly, during DMC cultivation, the vast majority of genes identified on the plasmids were down-regulated (79%), while the differentially expressed chromosomal genes were almost entirely up-regulated (93%). Global analysis of the upstream promoter regions of differentially expressed genes revealed that several share a common motif that may be important in transcriptional regulation during mammalian infection. Among genes with known or putative functions, the cell envelope category, which includes outer membrane proteins, was found to contain the most differentially expressed genes. The combined findings have generated a subset of genes that can now be further characterized to help define their role or roles with regard to B. burgdorferi virulence and Lyme disease pathogenesis.
Figures


References
-
- Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol. Microbiol. 18:507-520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

