Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection in mice
- PMID: 12761144
- PMCID: PMC155734
- DOI: 10.1128/IAI.71.6.3587-3596.2003
Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection in mice
Abstract
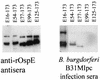
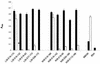
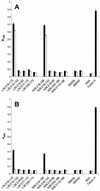
Immune evasion by Lyme spirochetes is a multifactorial process involving numerous mechanisms. The OspE protein family undergoes antigenic variation during infection and binds factor H (fH) and possibly FHL-1/reconectin. In Borrelia burgdorferi B31MI, the OspE family consists of three paralogs: BBL39 (ErpA), BBP38, and BBN38 (ErpP). BBL39 and BBP38 are identical and therefore are referred to here as BBL39. The goals of this study were to assess the specificity of the antibody (Ab) response to the OspE paralogs and to identify the domains or determinants of OspE that are required for the binding of fH and OspE-targeting Abs that develop during infection. Here we demonstrate that at least some of the anti-OspE Abs produced during infection are paralog specific and that Ab binding requires conformational determinants whose formation requires both the N- and C-terminal domains of OspE. The binding of fH to OspE was also found to be dependent on conformational determinants. It is also demonstrated here that all of the OspE paralogs expressed by B. burgdorferi B31MI are capable of binding fH. The binding of fH to members of the OspF protein family was also assessed. In contrast to an earlier report, no binding of BBO39 or BBR42 to human fH was detected. Lastly, a series of competitive binding enzyme-linked immunosorbent assay analyses, designed to determine if fH and infection serum Abs bind to the same sites on OspE, revealed that these ligands interact with different regions of OspE.
Figures




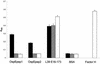
Similar articles
-
Demonstration of the involvement of outer surface protein E coiled coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H.J Immunol. 2004 Dec 15;173(12):7471-80. doi: 10.4049/jimmunol.173.12.7471. J Immunol. 2004. PMID: 15585873
-
Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis.Infect Immun. 2006 Mar;74(3):1967-72. doi: 10.1128/IAI.74.3.1967-1972.2006. Infect Immun. 2006. PMID: 16495576 Free PMC article.
-
Expression of complement factor H binding immunoevasion proteins in Borrelia garinii isolated from patients with neuroborreliosis.Eur J Immunol. 2005 Oct;35(10):3043-53. doi: 10.1002/eji.200526354. Eur J Immunol. 2005. PMID: 16208765
-
Complement evasion strategies of Borrelia burgdorferi sensu lato.FEBS Lett. 2020 Aug;594(16):2645-2656. doi: 10.1002/1873-3468.13894. Epub 2020 Aug 11. FEBS Lett. 2020. PMID: 32748966 Review.
-
Immune evasion of Borrelia burgdorferi: insufficient killing of the pathogens by complement and antibody.Int J Med Microbiol. 2002 Jun;291 Suppl 33:141-6. doi: 10.1016/s1438-4221(02)80027-3. Int J Med Microbiol. 2002. PMID: 12141738 Review.
Cited by
-
Illuminating the roles of the Borrelia burgdorferi adhesins.Trends Microbiol. 2013 Aug;21(8):372-9. doi: 10.1016/j.tim.2013.06.005. Epub 2013 Jul 19. Trends Microbiol. 2013. PMID: 23876218 Free PMC article. Review.
-
Serologic Evidence for the Exposure of Eastern Coyotes (Canis latrans) in Pennsylvania to the Tick-Borne Pathogens Borreliella burgdorferi and Anaplasma phagocytophilum.mSphere. 2020 Aug 12;5(4):e00544-20. doi: 10.1128/mSphere.00544-20. mSphere. 2020. PMID: 32817454 Free PMC article.
-
Evidence that the BBA68 protein (BbCRASP-1) of the Lyme disease spirochetes does not contribute to factor H-mediated immune evasion in humans and other animals.Infect Immun. 2006 May;74(5):3030-4. doi: 10.1128/IAI.74.5.3030-3034.2006. Infect Immun. 2006. PMID: 16622245 Free PMC article.
-
Lipoproteome screening of the Lyme disease agent identifies inhibitors of antibody-mediated complement killing.Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2117770119. doi: 10.1073/pnas.2117770119. Epub 2022 Mar 21. Proc Natl Acad Sci U S A. 2022. PMID: 35312359 Free PMC article.
-
Borrelia valaisiana resist complement-mediated killing independently of the recruitment of immune regulators and inactivation of complement components.PLoS One. 2013;8(1):e53659. doi: 10.1371/journal.pone.0053659. Epub 2013 Jan 8. PLoS One. 2013. PMID: 23320099 Free PMC article.
References
-
- Akins, D., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. - PubMed
-
- Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. - PubMed
-
- Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. - PubMed
-
- Cohen, G. H., V. J. Isola, J. Kuhns, P. W. Berman, and R. J. Eisenberg. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157-166. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

