Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum
- PMID: 12761390
- PMCID: PMC2323896
- DOI: 10.1110/ps.0300103
Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum
Abstract
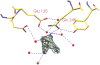
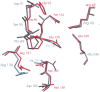
beta-1,4-Galactanases hydrolyze the galactan side chains that are part of the complex carbohydrate structure of the pectin. They are assigned to family 53 of the glycoside hydrolases and display significant variations in their pH and temperature optimum and stability. Two fungal beta-1,4-galactanases from Myceliophthora thermophila and Humicola insolens have been cloned and heterologously expressed, and the crystal structures of the gene products were determined. The structures are compared to the previously only known family 53 structure of the galactanase from Aspergillus aculeatus (AAGAL) showing approximately 56% identity. The M. thermophila and H. insolens galactanases are thermophilic enzymes and are most active at neutral to basic pH, whereas AAGAL is mesophilic and most active at acidic pH. The structure of the M. thermophila galactanase (MTGAL) was determined from crystals obtained with HEPES and TRIS buffers to 1.88 A and 2.14 A resolution, respectively. The structure of the H. insolens galactanase (HIGAL) was determined to 2.55 A resolution. The thermostability of MTGAL and HIGAL correlates with increase in the protein rigidity and electrostatic interactions, stabilization of the alpha-helices, and a tighter packing. An inspection of the active sites in the three enzymes identifies several amino acid substitutions that could explain the variation in pH optimum. Examination of the activity as a function of pH for the D182N mutant of AAGAL and the A90S/ H91D mutant of MTGAL showed that the difference in pH optimum between AAGAL and MTGAL is at least partially associated with differences in the nature of residues at positions 182, 90, and/or 91.
Figures




Similar articles
-
Effect of mutations on the thermostability of Aspergillus aculeatus β-1,4-galactanase.Comput Struct Biotechnol J. 2015 Apr 9;13:256-64. doi: 10.1016/j.csbj.2015.03.010. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 25941560 Free PMC article.
-
The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products.J Mol Biol. 2004 Jul 30;341(1):107-17. doi: 10.1016/j.jmb.2004.05.017. J Mol Biol. 2004. PMID: 15312766
-
Expression cloning, purification and characterization of a beta-1,4-galactanase from Aspergillus aculeatus.Curr Genet. 1995 Jan;27(2):135-41. doi: 10.1007/BF00313427. Curr Genet. 1995. PMID: 7788716
-
Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes.Prog Biophys Mol Biol. 2005 Nov;89(3):246-91. doi: 10.1016/j.pbiomolbio.2004.11.002. Epub 2004 Dec 29. Prog Biophys Mol Biol. 2005. PMID: 15950056 Review.
-
Structural and functional characterization of a family GH53 β-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.J Struct Biol. 2019 Jan 1;205(1):1-10. doi: 10.1016/j.jsb.2018.12.002. Epub 2018 Dec 13. J Struct Biol. 2019. PMID: 30553858 Review.
Cited by
-
Gene cloning, expression, and biochemical characterization of an alkali-tolerant β-mannanase from Humicola insolens Y1.J Ind Microbiol Biotechnol. 2012 Apr;39(4):547-55. doi: 10.1007/s10295-011-1067-8. Epub 2011 Dec 18. J Ind Microbiol Biotechnol. 2012. PMID: 22179540
-
Redesigning protein pKa values.Protein Sci. 2007 Feb;16(2):239-49. doi: 10.1110/ps.062538707. Epub 2006 Dec 22. Protein Sci. 2007. PMID: 17189477 Free PMC article.
-
Engineering the substrate binding site of the hyperthermostable archaeal endo-β-1,4-galactanase from Ignisphaera aggregans.Biotechnol Biofuels. 2021 Sep 16;14(1):183. doi: 10.1186/s13068-021-02025-6. Biotechnol Biofuels. 2021. PMID: 34530892 Free PMC article.
-
Genetic and biochemical characterization of a protease-resistant mesophilic β-mannanase from Streptomyces sp. S27.J Ind Microbiol Biotechnol. 2011 Mar;38(3):451-8. doi: 10.1007/s10295-010-0789-3. Epub 2010 Aug 5. J Ind Microbiol Biotechnol. 2011. PMID: 20686915
-
Structure of Aspergillus aculeatus β-1,4-galactanase in complex with galactobiose.Acta Crystallogr F Struct Biol Commun. 2019 Jun 1;75(Pt 6):399-404. doi: 10.1107/S2053230X19005612. Epub 2019 May 10. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31204685 Free PMC article.
References
-
- Aghajari, N., Feller, G., Gerday, C., and Haser, R. 1998. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 6 1503–1516. - PubMed
-
- Argos, P., Rossmann, M., Grau, U., Zuber, H., Frank, G., and Traschin, J. 1979. Thermal stability and protein structure. Biochemistry 25 5698–5703. - PubMed
-
- Braithwaite, K.L., Barna, T., Spurway, T.D., Charnock, S.J., Black, G.W., Hughes, N., Lakey, J.H., Virden, R., Hazlewood, G.P., Henrissat, B., et al. 1997. Evidence that galactanase A from Pseudomonas fluorescens subspecies cellulosa is a retaining family 53 glycosyl hydrolase in which E161 and E270 are the catalytic residues. Biochemistry 36 15489–15500. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. - PubMed
-
- Cerenius, Y., Ståhl, K., Svensson, A., Ursby, T., Oskarsson, Å., Albertsson, J., and Liljas, A. 2000. The crystallography beamline I711 at MAX II. J. Synchrotron Radiat. 7 203–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

