Why is Leu55-->Pro55 transthyretin variant the most amyloidogenic: insights from molecular dynamics simulations of transthyretin monomers
- PMID: 12761393
- PMCID: PMC2323890
- DOI: 10.1110/ps.0239703
Why is Leu55-->Pro55 transthyretin variant the most amyloidogenic: insights from molecular dynamics simulations of transthyretin monomers
Abstract
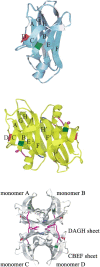
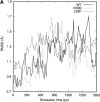
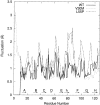
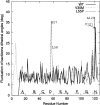
Transthyretin (TTR) is one of the known human amyloidogenic proteins. Its native state is a homotetramer with each monomer having a beta-sandwich structure. Strong experimental evidence suggests that TTR dissociates into monomeric intermediates and that the monomers subsequently self-assemble to form amyloid deposits and insoluble fibrils. However, details on the early steps along the pathway of TTR amyloid formation are unclear, although various experimental approaches with resolutions at the molecular or residue level have provided some clues. It is highly likely that the stability and flexibility of monomeric TTR play crucial roles in the early steps of amyloid formation; thereby, it is essential to characterize initial conformational changes of TTR monomers. In this article we probe the possibility that the differences in the monomeric forms of wild-type (WT) TTR and its variants are responsible for differential amyloidogenesis. We begin with the simulations of WT, Val30-->Met (V30M), and Leu55-->Pro (L55P) TTR monomers. Nanosecond time scale molecular dynamics simulations at 300 K were performed using AMBER. The results indicate that the L55P-TTR monomer undergoes substantial structural changes relative to fluctuations observed in the WT and V30M TTR monomers. The observation supports earlier speculation that the L55P mutation may lead to disruption of the beta-sheet structure through the disorder of the "edge strands" that might facilitate amyloidogenesis.
Figures



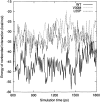
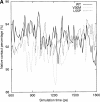

References
-
- Adams, D. and Said, G. 1996. Ultrastructural immunolabelling of amyloid fibrils in acquired and hereditary amyloid neuropathies. J. Neurol. 243 63–67. - PubMed
-
- Bashford, D. and Case, D.A. 2000. Generalized born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 51 129–152. - PubMed
-
- Batschelet, E. 1981. Circular statistics in biology. Academic Press, Inc., New York.
-
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81 3684–3690.
-
- Blake, C.C., Geisow, M.J., Oatley, S.J., Rerat, B., and Rerat, C. 1978. Structure of prealbumin: Secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 Å. J. Mol. Biol. 121 339–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

