Molecular characterization of the skate peripherin/rds gene: relationship to its orthologues and paralogues
- PMID: 12766040
- PMCID: PMC2991160
- DOI: 10.1167/iovs.02-1152
Molecular characterization of the skate peripherin/rds gene: relationship to its orthologues and paralogues
Abstract
Purpose: A great deal of information about functionally significant domains of a protein may be obtained by comparison of primary sequences of gene homologues over a broad phylogenetic base. This study was designed to identify evolutionarily conserved domains of the photoreceptor disc membrane protein peripherin/rds by analysis of the homologue in a primitive vertebrate, the skate.
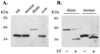
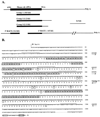
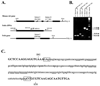
Methods: A skate retinal cDNA library was screened using a mouse peripherin/rds clone. The 5' and 3' untranslated regions of the skate peripherin/rds (srds) cDNA were isolated by the rapid amplification of cDNA ends (RACE) approach. The gene structure was characterized by PCR amplification and sequencing of genomic fragments. Northern and Western blot analyses were used to identify srds transcript and protein, respectively.
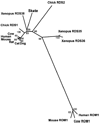
Results: A new homologue of peripherin/rds was identified from the skate retinal cDNA library. SRDS is a glycoprotein with a predicted molecular mass of 40.2 kDa. The srds gene consists of two exons and one small intron and transcribes into a single 6-kb message. Phylogenetic analysis places SRDS at the base of peripherin/rds family and near the division of that group and the branch leading to rds-like and rom-1 genes. SRDS protein is 54.5% identical with peripherin/rds across species. Identity is significantly higher (73%) in the intradiscal domains. Sequence comparison revealed the conservation of all residues that have been shown, on mutation, to associate with retinitis pigmentosa and showed conservation of most residues associated with macular dystrophies. Comparison with ROM-1 and other rds-like proteins revealed the presence of a highly conserved domain in the large intradiscal loop.
Conclusions: Srds represents the skate orthologue of mammalian peripherin/rds genes. Conservation of most of the residues associated with human retinal diseases indicates that these residues serve important functional roles. The high degree of conservation of a short stretch within the large intradiscal loop also suggests an important function for this domain.
Figures


Similar articles
-
Cysteine residues of photoreceptor peripherin/rds: role in subunit assembly and autosomal dominant retinitis pigmentosa.Biochemistry. 1998 Jan 13;37(2):680-5. doi: 10.1021/bi972036i. Biochemistry. 1998. PMID: 9425091
-
Structural and developmental analysis of the mouse peripherin/rds gene.Somat Cell Mol Genet. 1997 May;23(3):165-83. doi: 10.1007/BF02721369. Somat Cell Mol Genet. 1997. PMID: 9330629
-
Cloning of the human and murine ROM1 genes: genomic organization and sequence conservation.Hum Mol Genet. 1993 Apr;2(4):385-91. doi: 10.1093/hmg/2.4.385. Hum Mol Genet. 1993. PMID: 8504299
-
The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene.Prog Retin Eye Res. 2008 Mar;27(2):213-35. doi: 10.1016/j.preteyeres.2008.01.002. Epub 2008 Jan 26. Prog Retin Eye Res. 2008. PMID: 18328765 Review.
-
Role of peripherin/rds in vertebrate photoreceptor architecture and inherited retinal degenerations.Int Rev Cytol. 2006;253:131-75. doi: 10.1016/S0074-7696(06)53004-9. Int Rev Cytol. 2006. PMID: 17098056 Review.
Cited by
-
ROM1 is redundant to PRPH2 as a molecular building block of photoreceptor disc rims.bioRxiv [Preprint]. 2023 Aug 29:2023.07.02.547380. doi: 10.1101/2023.07.02.547380. bioRxiv. 2023. Update in: Elife. 2023 Nov 22;12:RP89444. doi: 10.7554/eLife.89444. PMID: 37693615 Free PMC article. Updated. Preprint.
-
A partial structural and functional rescue of a retinitis pigmentosa model with compacted DNA nanoparticles.PLoS One. 2009;4(4):e5290. doi: 10.1371/journal.pone.0005290. Epub 2009 Apr 24. PLoS One. 2009. PMID: 19390689 Free PMC article.
-
The Cys214-->Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice.Biochem J. 2005 Jun 1;388(Pt 2):605-13. doi: 10.1042/BJ20041960. Biochem J. 2005. PMID: 15656787 Free PMC article.
-
Modulating expression of peripherin/rds in transgenic mice: critical levels and the effect of overexpression.Invest Ophthalmol Vis Sci. 2004 Aug;45(8):2514-21. doi: 10.1167/iovs.04-0065. Invest Ophthalmol Vis Sci. 2004. PMID: 15277471 Free PMC article.
-
Structural and functional relationships between photoreceptor tetraspanins and other superfamily members.Cell Mol Life Sci. 2012 Apr;69(7):1035-47. doi: 10.1007/s00018-011-0736-0. Epub 2011 Jun 8. Cell Mol Life Sci. 2012. PMID: 21655915 Free PMC article. Review.
References
-
- Steinberg RH, Fisher SK, Anderson DH. Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol. 1980;190:501–518. - PubMed
-
- Boesze-Battaglia K, Lamba OP, Napoli AA, Jr, Sinha S, Guo Y. Fusion between retinal rod outer segment membranes and model membranes: a role for photoreceptor peripherin/rds. Biochemistry. 1998;37:9477–9487. - PubMed
-
- Ma J, Norton JC, Allen AC, et al. Retinal degeneration slow (rds) in mouse results from simple insertion of a t haplotype-specific element into protein-coding exon II. Genomics. 1995;28:212–219. - PubMed
-
- Cheng T, Al-Ubaidi MR, Naash MI. Structural and developmental analysis of the mouse peripherin/rds gene. Somat Cell Mol Genet. 1997;23:165–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

