Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles
- PMID: 12766229
- PMCID: PMC164679
- DOI: 10.1073/pnas.1232225100
Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles
Abstract
The MinCDE proteins of Escherichia coli are required for proper placement of the division septum at midcell. The site selection process requires the rapid oscillatory redistribution of the proteins from pole to pole. We report that the three Min proteins are organized into extended membrane-associated coiled structures that wind around the cell between the two poles. The pole-to-pole oscillation of the proteins reflects oscillatory changes in their distribution within the coiled structure. We also report that the E. coli MreB protein, which is required for maintaining the rod shape of the cell, also forms extended coiled structures, which are similar to the MreB structures that have previously been reported in Bacillus subtilis. The MreB and MinCDE coiled arrays do not appear identical. The results suggest that at least two functionally distinct cytoskeletal-like elements are present in E. coli and that structures of this type can undergo dynamic changes that play important roles in division site placement and possibly other aspects of the life of the cell.
Figures

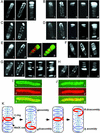
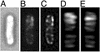

Comment in
-
Bacterial cell division spirals into control.Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7423-4. doi: 10.1073/pnas.1332806100. Epub 2003 Jun 16. Proc Natl Acad Sci U S A. 2003. PMID: 12810947 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

