Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants
- PMID: 12767980
- PMCID: PMC156182
- DOI: 10.1128/jvi.77.12.6601-6612.2003
Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants
Abstract
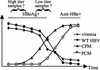
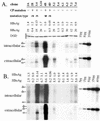
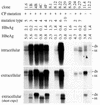
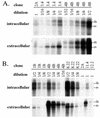
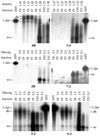
The core promoter mutants of hepatitis B virus (HBV) emerge as the dominant viral population at the late HBeAg and the anti-HBe stages of HBV infection, with the A1762T/G1764A substitutions as the hotspot mutations. The double core promoter mutations were found by many investigators to moderately enhance viral genome replication and reduce hepatitis B e antigen (HBeAg) expression. A much higher replication capacity was reported for a naturally occurring core promoter mutant implicated in the outbreak of fulminant hepatitis, which was caused by the neighboring C1766T/T1768A mutations instead. To systemically study the biological properties of naturally occurring core promoter mutants, we amplified full-length HBV genomes by PCR from sera of HBeAg(+) individuals infected with genotype A. All 12 HBV genomes derived from highly viremic sera (5 x 10(9) to 5.7 x 10(9) copies of viral genome/ml) harbored wild-type core promoter sequence, whereas 37 of 43 clones from low-viremia samples (0.2 x 10(7) to 4.6 x 10(7) copies/ml) were core promoter mutants. Of the 11 wild-type genomes and 14 core promoter mutants analyzed by transfection experiments in human hepatoma cell lines, 6 core promoter mutants but none of the wild-type genomes replicated at high levels. All had 1762/1764 mutations and an additional substitution at position 1753 (T to C), at position 1766 (C to T), or both. Moreover, these HBV clones varied greatly in their ability to secrete enveloped viral particles irrespective of the presence of core promoter mutations. High-replication clones with 1762/1764/1766 or 1753/1762/1764/1766 mutations expressed very low levels of HBeAg, whereas high-replication clones with 1753/1762/1764 triple mutations expressed high levels of HBeAg. Experiments with site-directed mutants revealed that both 1762/1764/1766 and 1753/1762/1764/1766 mutations conferred significantly higher viral replication and lower HBeAg expression than 1762/1764 mutations alone, whereas the 1753/1762/1764 triple mutant displayed only mild reduction in HBeAg expression similar to the 1762/1764 mutant. Thus, core promoter mutations other than those at positions 1762 and 1764 can have major impact on viral DNA replication and HBeAg expression.
Figures




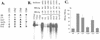
References
-
- Baptista, M., A. Kramvis, and M. Kew. 1999. High prevalence of 1762T 1764A mutations in the basic core promoter of hepatitis B virus isolated from Black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29:946-953. - PubMed
-
- Bowyer, S., L. van Staden, M. Kew, and J. Sim. 1997. A unique segment of the hepatitis B virus group A genotype identified in isolates from South Africa. J. Gen. Virol. 78:1719-1729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

