Stem-loop III in the 5' untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication
- PMID: 12767992
- PMCID: PMC156170
- DOI: 10.1128/jvi.77.12.6720-6730.2003
Stem-loop III in the 5' untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication
Abstract
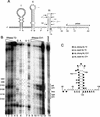
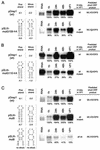
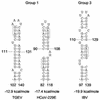
Higher-order structures in the 5' untranslated region (UTR) of plus-strand RNA viruses are known in many cases to function as cis-acting elements in RNA translation, replication, or transcription. Here we describe evidence supporting the structure and a cis-acting function in defective interfering (DI) RNA replication of stem-loop III, the third of four predicted higher-order structures mapping within the 210-nucleotide (nt) 5' UTR of the 32-kb bovine coronavirus (BCoV) genome. Stem-loop III maps at nt 97 through 116, has a calculated free energy of -9.1 kcal/mol in the positive strand and -3.0 kcal/mol in the negative strand, and has associated with it beginning at nt 100 an open reading frame (ORF) potentially encoding an 8-amino-acid peptide. Stem-loop III is presumed to function in the positive strand, but its strand of action has not been established. Stem-loop III (i) shows phylogenetic conservation among group 2 coronaviruses and appears to have a homolog in coronavirus groups 1 and 3, (ii) has in all coronaviruses for which sequence is known a closely associated short, AUG-initiated intra-5' UTR ORF, (iii) is supported by enzyme structure-probing evidence in BCoV RNA, (iv) must maintain stem integrity for DI RNA replication in BCoV DI RNA, and (v) shows a positive correlation between maintenance of the short ORF and maximal DI RNA accumulation in BCoV DI RNA. These results indicate that stem-loop III in the BCoV 5' UTR is a cis-acting element for DI RNA replication and that its associated intra-5' UTR ORF may function to enhance replication. It is postulated that these two elements function similarly in the virus genome.
Figures



Similar articles
-
Stem-loop IV in the 5' untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication.J Virol. 2005 Oct;79(19):12434-46. doi: 10.1128/JVI.79.19.12434-12446.2005. J Virol. 2005. PMID: 16160171 Free PMC article.
-
Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements.J Virol. 2009 Jun;83(12):6087-97. doi: 10.1128/JVI.00160-09. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357173 Free PMC article.
-
An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication.J Virol. 2007 Jul;81(14):7716-24. doi: 10.1128/JVI.00549-07. Epub 2007 May 2. J Virol. 2007. PMID: 17475638 Free PMC article.
-
The structure and functions of coronavirus genomic 3' and 5' ends.Virus Res. 2015 Aug 3;206:120-33. doi: 10.1016/j.virusres.2015.02.025. Epub 2015 Feb 28. Virus Res. 2015. PMID: 25736566 Free PMC article. Review.
-
Coronavirus genome structure and replication.Curr Top Microbiol Immunol. 2005;287:1-30. doi: 10.1007/3-540-26765-4_1. Curr Top Microbiol Immunol. 2005. PMID: 15609507 Free PMC article. Review.
Cited by
-
A 5'-proximal stem-loop structure of 5' untranslated region of porcine reproductive and respiratory syndrome virus genome is key for virus replication.Virol J. 2011 Apr 15;8:172. doi: 10.1186/1743-422X-8-172. Virol J. 2011. PMID: 21496223 Free PMC article.
-
Dependence of coronavirus RNA replication on an NH2-terminal partial nonstructural protein 1 in cis.J Virol. 2014 Aug;88(16):8868-82. doi: 10.1128/JVI.00738-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872586 Free PMC article.
-
Characterization of Chinese Porcine Epidemic Diarrhea Virus with Novel Insertions and Deletions in Genome.Sci Rep. 2017 Mar 9;7:44209. doi: 10.1038/srep44209. Sci Rep. 2017. PMID: 28276526 Free PMC article.
-
De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures.Nucleic Acids Res. 2021 Apr 6;49(6):3092-3108. doi: 10.1093/nar/gkab119. Nucleic Acids Res. 2021. PMID: 33693814 Free PMC article.
-
Putative cis-acting stem-loops in the 5' untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts.J Virol. 2006 Nov;80(21):10600-14. doi: 10.1128/JVI.00455-06. Epub 2006 Aug 18. J Virol. 2006. PMID: 16920822 Free PMC article.
References
-
- Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

